Classification of brain tumor type and grade using mri texture and shape in a machine learning scheme
Magnetic Resonance in Medicine 62:1609 –1618 (2009)
Classification of Brain Tumor Type and Grade Using MRI
Texture and Shape in a Machine Learning Scheme
Evangelia I. Zacharaki,1,2* Sumei Wang,1 Sanjeev Chawla,1 Dong Soo Yoo,1,3Ronald Wolf,1 Elias R. Melhem,1 and Christos Davatzikos1
The objective of this study is to investigate the use of pattern
lowing surgical biopsy or resection, but this also has lim-
classification methods for distinguishing different types of brain
itations, including sampling error and variability in
tumors, such as primary gliomas from metastases, and also for
grading of gliomas. The availability of an automated computer
The objective of this study is to provide an automated
analysis tool that is more objective than human readers can
tool that may assist in the imaging evaluation of brain
potentially lead to more reliable and reproducible brain tumor
neoplasms by determining the glioma grade and differen-
diagnostic procedures. A computer-assisted classification
method combining conventional MRI and perfusion MRI is de-
tiating between different tissue types, such as primary
veloped and used for differential diagnosis. The proposed
neoplasms (gliomas) from secondary neoplasms (metasta-
scheme consists of several steps including region-of-interest
ses). These issues are of critical clinical importance in
definition, feature extraction, feature selection, and classifica-
making decisions regarding initial and evolving treatment
tion. The extracted features include tumor shape and intensity
strategies, and conventional MR imaging is often not ade-
characteristics, as well as rotation invariant texture features.
quate in providing answers (1,5). Automated tools, if
Feature subset selection is performed using support vector
proven accurate, can ultimately be applied to (i) provide
machines with recursive feature elimination. The method was
more reliable differentiation, especially when the neo-
applied on a population of 102 brain tumors histologically diag-
plasm is heterogeneous and therefore cannot be ade-
nosed as metastasis (24), meningiomas (4), gliomas World
quately sampled by localized needle biopsy; (ii) avoid
Health Organization grade II (22), gliomas World Health Orga-
nization grade III (18), and glioblastomas (34). The binary sup-
invasive procedures such as biopsy, especially in cases
port vector machine classification accuracy, sensitivity, and
where the risks outweigh the benefits; and (iii) expedite or
specificity, assessed by leave-one-out cross-validation, were,
anticipate the diagnosis (histologic examination is usually
respectively, 85%, 87%, and 79% for discrimination of metas-
time consuming).
tases from gliomas and 88%, 85%, and 96% for discrimination
Toward a similar goal, researchers used conventional
of high-grade (grades III and IV) from low-grade (grade II) neo-
MR imaging and echo-planar relative cerebral blood vol-
plasms. Multiclass classification was also performed via a one-
ume (rCBV) maps calculated from perfusion imaging to
vs-all voting scheme.
Magn Reson Med 62:1609 –1618, 2009.
differentiate between high-grade and low-grade neoplasms
2009 Wiley-Liss, Inc.
(6) or assessed the contribution of MR perfusion alone in
Key words: brain tumor; MRI; classification; SVM; feature se-
differentiating certain tumor types (7,8). Many studies
lection; texture; tumor grade
have used MR spectroscopy for brain tumor classification
Clinical decisions regarding the treatment of brain neo-
(9-11). Specifically, spectroscopic and conventional MR
plasms rely, in part, on MRI at various stages of the treat-
imaging was used in Wang et al. (9) to differentiate benign
ment process. Radiologic diagnosis is based on the mul-
from malignant brain neoplasms, applying a decision tree
tiparametric imaging profile (CT, conventional MRI, ad-
algorithm, whereas spectroscopic and perfusion MRI was
vanced MRI). Tumor characterization is difficult because
used in Weber et al. (10) to evaluate the inherent hetero-
neoplastic tissue is often heterogeneous in spatial and
geneity of brain neoplasms by defining four regions of
imaging profiles (1), and for some imaging techniques of-
interest (ROIs) in the tumoral and peritumoral region.
ten overlaps with normal tissue (especially the infiltrating
Some studies used apparent diffusion coefficient maps
part) (2,3). Gliomas might show mixed characteristics; for
computed from diffusion tensor imaging data to differen-
example, demonstrating both low- and high-grade fea-
tiate metastases from primary cerebral tumors (by measur-
tures. The reference standard for characterizing brain neo-
ing diffusion in peritumoral edema) (12) or combined ap-
plasms is currently based on histopathologic analysis fol-
parent diffusion coefficient with rCBV to differentiate tu-mefactive demyelinating lesions and primary neoplasmsfrom abscesses and lymphomas (13).
The previous studies are very useful in determining the
1Department of Radiology, University of Pennsylvania, Philadelphia, Pennsyl-vania, USA
clinical significance of each MR sequence separately; how-
2Laboratory of Medical Physics, School of Medicine, University of Patras, Rio,
ever, they do not investigate nonlinear relationships be-
tween different variables by pattern analysis. Pattern clas-
3Department of Radiology, Dankook University Hospital, Chungchungnam-
sification techniques were applied for differentiating brain
neoplasms based on linear discriminant analysis (LDA)
*Correspondence to: Evangelia Zacharaki, PhD, Department of Radiology,University of Pennsylvania, 3600 Market St, Philadelphia, PA 19104. E-mail
(14,15), or independent component analysis (16) on spec-
tral intensities. Others applied support vector machines
Received 9 March 2009; revised 23 June 2009; accepted 24 June 2009.
(SVMs) on perfusion MRI (17) or combined variable selec-
DOI 10.1002/mrm.22147
tion and classification using bayesian least squares SVMs
Published online 26 October 2009 in Wiley InterScience (www.interscience.
wiley.com).
and relevance vector machines on microarray or spectros-
2009 Wiley-Liss, Inc.
Zacharaki et al.
copy data (18). Textural features from
T1 postcontrast im-
used to test the robustness and accuracy of the proposed
ages were employed in (19) to discriminate between met-
astatic and primary brain tumors using a probabilisticneural network with a nonlinear least squares featurestransformation method. Although these studies apply
MATERIALS AND METHODS
more advanced machine learning techniques, they use a
We propose a multiparametric framework for brain tumor
single MR sequence and do not investigate the contribu-
classification and prediction of degree of malignancy. In-
tion of multiple imaging parameters. Multiparametric fea-
tensity- and texture-based features are integrated via a
tures are explored by nonlinear classification techniques
pattern classification technique into a multiparametric im-
in (20,21). Li et al. (20) classify gliomas according to their
aging profile. The features are first normalized to have zero
clinical grade using linear SVMs trained on a maximum of
mean and unit variance. A feature selection method is then
15 descriptive features (such as amount of mass effect or
used to select a small set of effective features for classifi-
blood supply), which are estimated quantitatively by do-
cation in order to improve the generalization ability and
main experts. The definition of such features is based on
the performance of the classifier.
expert knowledge and therefore is not completely auto-
In this section, the clinical data and the acquisition
mated and reproducible. Devos et al. (21) combine stan-
protocol are first reviewed. Then, the preprocessing of the
dard MR intensities with spectroscopy imaging to improve
data and the ROIs for feature extraction are described,
classification performance using three classification tech-
followed by the definition of the features. Finally, the
methods for feature selection and classification are pre-
In this study, we explore the heterogeneous regions of
brain tumors by combining imaging features from severalsequences and extract morphologic and texture character-
istics. Our analysis requires three ROIs, which define theneoplastic and necrotic region on contrast enhanced
T
We examined 98 patients (52 women, 46 men; age 17-83
weighted MRI, and edematous region on fluid attenuated
years) with a diagnosis of brain neoplasm (from September
inversion recovery (FLAIR) image. Instead of only measur-
2006 to December 2007) who had not been treated at the
ing mean values in the ROIs, we investigate if conven-
time of MRI. Four patients had multiple (2), not related to
tional MRI has higher potential when complicated features
each other, lesions that were regarded as independent
are extracted, such as rotation invariant texture features
masses. All patients underwent biopsy or surgical resec-
based on Gabor filtering (22). Moreover, since it has been
tion of the tumor with histopathological diagnosis. The
shown that contrast enhancement in conventional MRI is
total of 102 brain masses were histologically diagnosed
not sufficient for glioma grading or differentiating between
and graded based on World Health Organization criteria as
metastasis and high grade tumor, we also incorporate
rCBV maps. This approach incorporates imaging data
which are (or can be) acquired in a routine clinical proto-
gliomas grade II (22) including astrocytomas, oligo-
col, including multiparametric conventional MRI and per-
and gliomatosis cerebri
We apply the method for binary classification, but also
gliomas grade III (18) including anaplastic astrocyto-
investigate the multiclass classification problem for differ-
mas and (anaplastic) oligodendrogliomas
entiating between the most common brain tumors: metas-
glioblastomas (GBMs) (34) including 1 giant cell GBM
tasis, meningioma (usually grade I), low-grade glioma(grade II), grade III glioma, and glioblastoma (grade IV)
The primary sites of cancer for patients with metastatic
according to the World Health Organization system. Grade
lesions were lung (14), breast (5), melanoma (3), and
II and grade III gliomas include astrocytomas (anaplastic or
esophagus (1). For one case, the primary tumor type could
not), oligodendrogliomas, and oligoastrocytomas. The pro-
not be confirmed but most likely came from breast or lung.
posed framework consists of a training step, where the
The study was approved by the institutional review board
classifier learns the imaging characteristics of the different
and was compliant with the Health Insurance Portability
tumor types, and a testing step, where the classifier recog-
and Accountability Act.
nizes the tumor type in a new image. The framework is
The patients were imaged using a 3.0-T MRI scanner
general and, if a significant number of observations (train-
system (Magnetom Trio Tim System; Siemens Medical
ing samples) exist, can be applied to classify any other
Systems, Erlangen, Germany), with a multichannel
neoplasm and also non-neoplastic pathologies, including
phased-array coil. The imaging acquisition protocol was
lymphoma, abscess, and encephalitis. We differentiate be-
the same for all patients and includes the following se-
tween tumor types by combining multiparametric MR im-
quences: axial three-dimensional
T1-weighted (
T1) (mag-
ages into a single classification rule rather than using sin-
netization prepared rapid acquisition gradient echo pulse
gle modalities independently. We exploit the potential of
repetition time/echo time/inversion time 1760/3.1/950,
features extracted automatically from the images in order
matrix size 192 ⫻ 256, pixel spacing 0.9766 ⫻ 0.9766 mm,
to avoid descriptive criteria, which are rater dependent
slice thickness 1 mm), sagittal three-dimensional
T2-
and require prior knowledge (the help of experts).
weighted (
T2) (matrix size 256 ⫻ 320, pixel spacing
The proposed scheme consists of four parts: ROI defini-
0.8969 ⫻ 0.8969 mm, slice thickness 0.9 mm), FLAIR
tion, feature extraction, feature selection, and classifica-
(pulse repetition time/echo time/inversion time 9420/141/
tion based on SVMs. Leave-one-out cross-validation is
2500, matrix size 192 ⫻ 256, pixel spacing 0.9375 ⫻
Tumor Classification Using Machine Learning
0.9375 mm, slice thickness 3 mm) and diffusion tensor
Feature Description
imaging. Diffusion tensor imaging was not used as part of
We chose a large number of features (161) for investiga-
this study but will be incorporated in the future. Axial
tion, which included age, tumor shape characteristics, im-
three-dimensional contrast-enhanced
T1-weighted images
age intensity characteristics within some of the ROIs, and
(T1ce) were obtained after administration of a standard
Gabor texture features, as explained next.
dose (0.1 mmol/kg) of gadodiamide with a power injector(Medrad, Idianola, PA). Also, after an initial loading doseof 3 mL gadodiamide and a 5-min delay,
T
Shape and Statistical Characteristics of Tumor (Evaluated
in ROI1 艛
ROI2 艛
ROI3)
dynamic susceptibility perfusion MRI was performed us-ing a gradient echo EPI acquisition during bolus injection
Five shape features (
si,
i ⫽ 1, . . , 5) of the total tumor area
of 12 mL gadodiamide. Twenty slices were acquired with
are investigated, i.e., the tumor circularity, irregularity,
pulse repetition time/echo time 2000/45 ms, matrix size
rectangularity, the entropy of radial length distribution of
128 ⫻ 128, pixel spacing 1.7 ⫻ 1.7 mm, slice thickness
the boundary voxels, and the surface-to-volume ratio. Also
3.0 mm. rCBV maps were generated off-line by calculating
three statistical features are calculated, i.e., the ratio of
the change in relaxation rate using the equation ⌬
R2*(
t) ⫽
enhancing (
venh), necrotic (
vnec), and edematous (
vedm) tu-
⫺ln(
S(t)/S0)/TE, where
S(
t) is signal intensity at time
t and
mor volume vs total (enhancing and nonenhancing) tumor
S0 is baseline signal intensity, and then integrating under
the ⌬
R2*(
t) curve over time points corresponding to thefirst pass of the contrast bolus.
Image Intensity Characteristics
The mean () and variance (
var) of image intensities of
T
Preprocessing and Definition of ROIs
T1ce,
T2 are calculated in the central and marginal area of
The images are preprocessed following a number of steps,
ROI1, ROI2 and ROI3. For FLAIR images, the same inten-
including noise reduction, bias-field correction, and rigid
sity characteristics are extracted from ROI4. For the rCBV
intrasubject registration using the FSL library of analysis
maps, since hyperintense areas are indicative but not spe-
tools (23). The coregistration of all sequences (
T1, T1ce,
T2,
cific to tumor, we first mask out areas that appear hypoin-
FLAIR, rCBV) is required in order to extract features from
tense in FLAIR and then calculate the mean and variance
the ROIs and is performed with the rigid registration algo-
of rCBV in the central and marginal region of ROI1, ROI2,
rithm FLIRT (24) from FSL. The intensity levels are made
and ROI4. More analytically, the voxels with low intensity
comparable across subjects by histogram matching. For
on the FLAIR image, such as ventricles or peripheral ves-
this purpose skull stripping is first performed using brain
sels in the cortical sulci, are excluded from the analysis of
extraction tool (BET) (25) to generate a brain tissue mask
the rCBV maps because they may represent normal vascu-
from the
T1 image, which is then used to extract the brain
lature that may be indistinguishable from abnormal neo-
region from all other coregistered sequences. A linear
vascularity due to neoplastic infiltration. All intensity-
transformation of the intensities (translation and scaling)
related features sum up to 52 features in total. These fea-
is applied in order to minimize the
L
2-norm of the histo-
tures are denoted as
C I兲,
varC I兲, when measured in the
gram difference between each subject and a template im-
central area and
R 共
m I兲,
varm I兲, when measured in the mar-
age. Histogram matching is not applied to the rCBV maps,
ginal area, where
R 僆 {1, . . , 4} for ROI1 to ROI4, and
I 僆
calculated from the perfusion sequence.
{
T1, T1ce,
T2, FLAIR, rCBV}.
The features were extracted from ROIs manually traced
by two expert neuroradiologists. A maximum of four ROIs
Gabor Texture
The voxel-wise texture features of image
I(x,y,z) are ex-
ROI1 (neoplastic, enhancing), ROI2 (neoplastic, non-
tracted at each tomographic slice of the three-dimensional
enhancing): includes all non-necrotic enhancing neo-
ROI by convoluting with 2D Gabor filters (26,27) and av-
plastic tissue, or, if the lesion did not show enhance-
eraging inside the ROI. The 2D Gabor filters are mathemat-
ment, the whole non-necrotic
T1-hypointense neo-
ically described at location (x,y) as
plastic tissue, avoiding peritumoral edema by tracingthe FLAIR image.
⫹ ␥2
y
ROI3 (necrotic): this ROI is delineated only in cases
g,,,共
x,
y兲 ⫽ exp冉 ⫺
x ⫹ 冊
including necrotic tumor tissue by tracing the coreg-istered T1ce,
T1,
T2, and FLAIR images and by exclud-
ing hemorrhage.
ROI4 (edematous): FLAIR and
T2 images are used to
depict the peritumoral edema (possibly including
⫽
xcos共兲 ⫹
ysin共兲
and y ⫽ ⫺
xsin共兲 ⫹
ycos共兲
neoplastic infiltration), drawing the ROI surroundingthe high signal intensity seen on these sequences.
and ⫽ 1/
f is the wavelength, the orientation, ␥ thespatial aspect ratio that determines the eccentricity of the
It should be noted that the ROIs are drawn based on the
convolution kernel and which was taken as ␥ ⫽ 1 in this
signal intensity of different sequences and are not ex-
study, and the phase offset that determines the symmetry
pected to be highly specific to each tissue type. For exam-
of the Gabor function. The standard deviation of the
ple, nonenhancing neoplastic tissue and edematous tissue
gaussian factor determines the neighborhood of a voxel in
might overlap and be both present in ROI2 or ROI4.
which weighted summation takes place. The ratio /
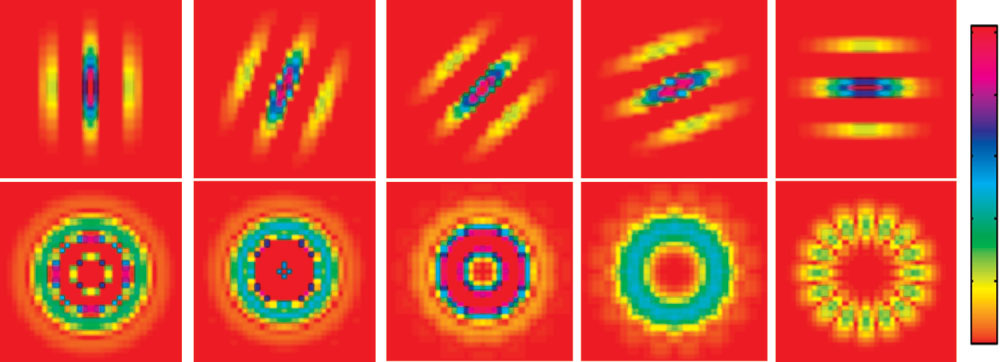
Zacharaki et al.
FIG. 1. Examples of filters usedto extract texture features. The1st row shows Gabor filters forsame frequency and different ori-entations, and the 2nd row, therotation-invariant filters.
determines the spatial frequency bandwidth. We used a
Therefore for each bandwidth we obtain 25 rotation-in-
constant ratio / (27), and therefore is not considered as
variant texture features, gc共
I I兲, where l ⫽ 1, . . , 5 is the
an independent parameter.
wavelength index and c ⫽ 1, . . , 5 is the index on fast
We calculated the texture by combining the output of a
Fourier transform coefficients and I 僆 { T1ce, FLAIR }. We
symmetric ( ⫽ 0) and antisymmetric ( ⫽ /2) Gabor
used two bandwidth values (1 and 1.5) and finally ob-
kernel using the L2-norm (27), as shown below:
tained 100 features in total describing texture in T1ce andFLAIR for tumor classification.
Figure 1 illustrates in the first row the Gabor filter for
a single frequency across the first five (out of eight)
I共x,y兲 丢 g
,,⫽0 x,y兲兲2 ⫹ 共I共x,y兲 丢 g,,⫽/2 x,y兲兲2
orientations and in the 2nd row the rotation-invariant
where 丢 denotes 2D linear convolution. Then, in order to
filters after fast Fourier transform for the same fre-
make the average Gabor features rotation invariant, for
quency. Figure 2 illustrates examples of brain tumor
each radial frequency f fast Fourier transform is performed
types and the corresponding texture images. The first
across orientation (22). Suppose we use N orientations
row shows one axial slice of the T1ce image with the
within a period of , then N magnitudes of Fourier coef-
tumoral region of interest (ROI1 艛 ROI2 艛 ROI3) indi-
ficients can be obtained for each frequency f, but only the
cated by a red line. The 2nd row shows the same slice in
FLAIR image zoomed around the tumor region. As an
c ⫽ N/2 ⫹ 1 coefficients for each frequency are
example of texture, one pattern extracted from FLAIR is
We extract the texture features from the T1ce in ROI1 艛
shown over the edematous area (ROI4). The illustration
ROI2 艛 ROI3 and from FLAIR in ROI4. We use five wave-
shows the voxelwise texture without averaging over the
lengths, ⫽ 兵2冑2,4,4冑2,8,8冑2其, and N ⫽ 8 orientations
area of interest.
in [0, ] which after the fast Fourier transform produce
It should be noted that texture features are affected by
Nc ⫽ N/2 ⫹ 1 ⫽ 5 unique coefficients for each frequency.
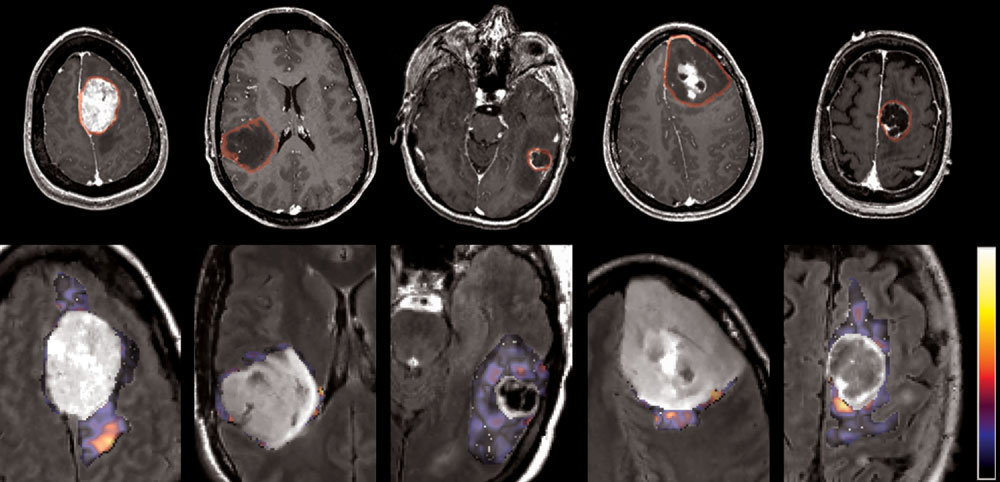
the MRI acquisition parameters, especially the spatial res-
FIG. 2. MR images of different of brain tumor types and an example of texture images extracted from the edematous area. From left to right:meningioma, glioma grade II, grade III, grade IV, and metastasis. First row: T1ce image with the tumoral region of interest. Second row:FLAIR image (zoomed in the tumor region) overlaid with one of the textural patterns ( ⫽ 8). This pattern is shown here as voxelwise texturefor illustration purposes and is not equivalent to our calculations. The average texture values (calculated before fast Fourier transform)(feature g54) proved to be significant in discrimination of meningiomas.
Tumor Classification Using Machine Learning
olution (28). Therefore it is important to use the same
graph. The remaining features (with P value ⬎ ␣) are
acquisition protocol during training of the classifier and
ranked by applying the t test using a bagging strategy (31).
during testing, i.e., when the classifier learns the imaging
Specifically, the training set is divided in five folds and the
characteristics and when the classifier recognizes a new
features are ranked at each fold separately. Then the rank-
case, respectively.
ings are combined to produce the total rank for each fea-ture based on forward selection.
Feature Selection
Feature Subset Selection Method Based on SVM
Feature selection methods can be divided into featureranking methods and feature subset selection methods
SVM-based feature selection methods have been success-
(29). The feature ranking methods compute a ranking score
fully applied in a variety of problems. One good example
for each feature according to its discriminative power and
is the support vector machine recursive feature elimina-
then simply select the top ranked features as final features
tion (SVM-RFE) algorithm, which was initially proposed
for classification. These feature selection methods are pref-
for a cancer classification problem (32) and was later ex-
erable for high dimensional problems due to their compu-
tended by introducing SVM-based leave-one-out error
tational scalability. However, the subset of features se-
bound criteria in (33). The goal of SVM-RFE is to find a
lected by the feature ranking methods might contain a lot
subset of features that optimize the performance of the
of redundant features since the ranking score is computed
classifier. This algorithm determines the ranking of the
independently for each feature, by completely ignoring its
features based on a backward sequential selection method
correlation with others. On the contrary, the feature subset
that removes one feature at a time. At each time, the
selection methods focus on selecting a subset of features
removed feature makes the variation of SVM-based leave-
that jointly have better discriminative power. In general,
one-out error bound smallest, compared to removing other
sophisticated subset selection methods have better classi-
fication performance than feature ranking methods, but
The training data set 兵 x
其 僆 ᑬn ⫻ 兵 ⫺
1,1其 consists of
their high computational cost usually limits their applica-
the training patterns xk and the known class labels yk,
tions to the high dimensional problems. Since combinato-
where k ⫽ 1, . . , NS (NS ⫽ N1 ⫹ N2 is the total number of
rial search of the optimal combination of features would be
training samples belonging to both classes). We apply the
computationally prohibitive, we combine the advantages
zero-order method for identifying the variable that pro-
of both the feature ranking method and the feature subset
duces the smallest value of the ranking criterion when
selection method. Specifically, we first reduce the number
removed, and use the weight magnitude 储w储2 as ranking
of features by eliminating the less relevant features using a
criterion, defined as
forward selection method based on a ranking criterion andthen apply backward feature elimination using a feature
subset selection method, as explained next.
where K(i) is the Gram matrix of the training data (see Eq. 2)
We use a simple ranking-based feature selection criterion,
when the variable i is removed and a共i兲 is the correspond-
a two-tailed t test, which measures the significance of a
ing solution of the SVM classifier.
difference of means between two distributions (30), andtherefore evaluates the discriminative power of each indi-
Constrained LDA
vidual feature in separating two classes. If the features areassumed to come from normal distributions with un-
For the purpose of comparison of the previously described
known but equal variances, the t statistic is defined as:
feature selection algorithm with a commonly used dimen-sionality reduction method, such as LDA, we also investi-
gated the performance of the constrained LDA algorithm
t ⫽ 冑共
for feature selection (34). Constrained LDA maximizes the
1兲v2冉 1 1
discriminant capability between classes without trans-
forming the original features, as done by traditional LDAor principal component analysis. This is important be-
where x1, v1 and x2, v2 are the sample mean and sample
cause it allows preservation of the physical meaning of the
variance of a particular feature in 1st and 2nd class, re-
input variables and assessment of the contribution of each
spectively. N1 and N2 are the number of samples in each
original variable in classification.
Since the correlation among features has been com-
Calculating Total Rank by Combination
pletely ignored in this feature ranking method, redundantfeatures are inevitably selected, which ultimately affects
Classification is performed by following a leave-one-out
the classification results. Therefore, we use this feature
strategy on the training samples. For each leave-one-out
ranking method to select the more discriminative features,
experiment, feature ranking is performed using data only
e.g., by applying a cutoff ratio (P value ⬍ ⫽ ␣, where ␣ ⫽
from the training samples. The feature selection method is
0.1 is the significance level for rejecting the null hypoth-
implemented in each training subset in order to correct for
esis), and then apply the feature subset selection method
the selection bias (35). It is important that cross-validation
on the reduced feature space, as detailed in the next para-
be external to the feature selection process in order to more
Zacharaki et al.
accurately estimate the prediction error. Evidently, there is
constant. The aim was to include a constant fraction of the
no guarantee that the same subset of features will be se-
training samples in a ball in feature space of size s for any
lected at each leave-one-out experiment. We combine the
dimensionality (number of features). The constant k was
rankings of all leave-one-out experiments and report the
determined such that the fraction of the training samples
total rank of features (in Table 2) according to the fre-
contained in the kernel is approximately 20%.
quency of a feature appearing in a specific rank. For ex-
The multiclass problem was solved by constructing and
ample the top-ranked feature is assumed to be the one that
combining several binary classifiers into a voting scheme.
more frequently has the highest ranking score, regardless
We applied majority voting from all one-vs-all binary clas-
of the distribution of the scores it receives across experi-
sification problems. For assessing the predictive ability of
ments. In order to assess if the features are selected con-
the classification scheme, we applied leave-one-out cross-
sistently, we use two measures: the entropy (E) to evaluate
validation. In the future, when more training data will be
the certainty in ranking and the standard deviation (S) to
available for each class, we will assess the generalization
measure the compactness. These measures are normalized
ability through 3-fold cross-validation and will further
between 0 (no consistency) and 1 (same rank in all exper-
optimize the classification parameters C and s for each
classification problem by exhaustive search.
Classification was performed by starting with the more
We applied leave-one-out external cross-validation in
discriminative features and gradually adding less discrim-
classifying meningioma (MEN), glioma of grades II, III,
inative features, until classification performance no longer
and IV (GL2, GL3, and GL4, respectively), and metasta-
improved. Three pattern classification methods were in-
sis (MET) by applying three different classification
vestigated for comparison: LDA with Fisher's discriminant
methods (LDA, kNN, nonlinear SVM) and two feature
rule (36), k-nearest neighbor (k-NN) (37), and nonlinear
ranking methods (t test with bagging, constrained LDA).
SVMs (33). In LDA, a transformation function is sought
The results are presented in Table 1. The first column in
that maximizes the ratio of between-class variance to with-
Table 1 shows the tumor type to be classified. The other
in-class variance. Since usually there is no transformation
columns show the number of selected features (NF) giv-
that provides complete separation, the goal is to find the
ing the highest classification accuracy (the feature selec-
transformation that minimizes the overlap of the trans-
tion step was cross-validated; however, we currently
formed distributions. k-NN neighbor classification is per-
have no automated way of selecting the number of fea-
formed based on closest training examples in the feature
tures; hence, we recorded performance as a function of
space. In SVMs, the original input space is mapped into a
the number of features), the classification accuracy
higher dimensional feature space in which an optimal
(ACC), defined as the percentage of correctly classified
separating hyperplane is constructed such that the dis-
samples, and the area under the receiver operating char-
tance from the hyperplane to the nearest data point is
acteristic curve (AUC). The last row shows the corre-
maximized. Due to this property, SVM classifiers tend to
sponding mean values. The results show that the clas-
possess good generalization ability.
sification accuracy is higher when using SVM, as ex-
A critical parameter in SVM is the penalty parameter C.
pected. The number of features giving highest accuracy
A larger C corresponds to assigning a higher penalty to
is overall smaller when t test is used as ranking criterion
misclassification. Since the data are unbalanced and the
vs constrained LDA.
sample size is rather small to produce balanced classes by
Subsequently, we investigated the performance of SVM
subsampling the largest class, we used a weighted SVM1
using the SVM-RFE algorithm for feature selection and
(38) to apply larger penalty to the class with the smaller
assessed the contribution of each feature in classification.
number of samples. If the penalty parameter is not
The results are shown in Table 2. The first 10 rows show
weighted (equal C for both classes), there is an undesirable
the classification between two single tumor types, similar
bias toward the class with the large training size; thus we
to Table 1, whereas the last two rows show the classifica-
set the ratio of penalties for the two classes, C1 and C2, (in
tion between combined types: secondary vs primary glio-
each binary classification), to the inverse ratio of the train-
mas (i.e., metastases vs gliomas grades II, III, IV) and low
ing class sizes (38). The kernel function used in our SVM
vs high grade gliomas (grade II vs III and IV). Those two
classifier is gaussian radial basis function kernel. The
classification problems are clinically relevant since treat-
Gram matrix for two feature vectors xi,xj is defined as
ment is usually adapted accordingly. Meningiomas are notincluded in the combined classification problems because
储x ⫺ 储2
they differ from the glial tumors and metastases in both
origin and behavior.
Table 2 also shows the top-ranked features (thresh-
where s controls the size of the gaussian kernel. We de-
olded based on t statistic and ranked by the SVM-RFE
fined s to be adaptive to the number of retained features
algorithm for each classification task. Not all selected
(NF), using the equation s ⫽ k 䡠 N 䡠
log共NF , where k is a
features are shown in the third column, but only theoverall top ranked calculated by combining all leave-one-out results. The notation used for these features is
described analytically in the Feature Description sec-
Chang C-C, Lin C-J. LIBSVM: a library for support vector machines, 2001.
Software available at http://www.csie.ntu.edu.tw/⬃cjlin/libsvm.
tion. The subsequent columns show the sensitivity,
Tumor Classification Using Machine Learning
Table 1Binary Classification Accuracy (Acc) and AUC Obtained by Leave-One-Out Cross-Validation Using Different Classifiers (LDA, k-NN,SVM) and Feature Ranking Methods (t Test With Bagging or CLDA)
k-NN (k ⫽ 3)
specificity, accuracy, and area under the receiver oper-
be a relevant pattern. It is interesting to note that usually
ating characteristic values, respectively.
the large wavelengths ( ⫽ 8 or 8冑2) appear as part of
The results of Table 2 show that perfusion is an impor-
the selected patterns.
tant MR imaging technique for most classification tasks.
For most binary classification pairs, the number of fea-
The enhancing portion in T1ce appears as significant pa-
tures selected with SVM-RFE giving the highest classifica-
rameter in the distinction of meningioma from gliomas of
tion accuracy is relatively small, which means that the
all grades; specifically in the case of glioblastoma, the
chance of overfitting is reduced and the generalization
enhancing portion in T1ce is selected as a single feature
ability improved. The feature reduction is an important
and achieves 97.4% accuracy. A combination of multipa-
advantage of SVM-RFE over the simple t test. Although the
rametric features, including T2 and T1 precontrast imaging
accuracy over all classification tasks is on average almost
characteristics, is used for classifying primary glioma vs
the same when applying SVM-RFE as a second step after t
metastasis (with accuracy 84.7%) and low- vs high-grade
test, the average number of selected features over all clas-
glioma (with accuracy 87.8%). Gabor texture seems also to
sification tasks is overall reduced by 33% (N
Table 2Binary Classification Results Obtained by Leave-One-Out Cross-Validation Using SVM for Classification and SVM-RFE for FeatureSelection
Overall top-ranked features (maximum eight
features are shown)
enh, varm(rCBV), c(T1), s3, m(T2),
5(FLAIR), venh, vnec, c(T1)
Zacharaki et al.
Table 3Multiclass Confusion Matrix Obtained With One-Versus-All VotingScheme Using SVM Classifiers With RFE
Prediction, NF ⫽ 18 (out of 50)
and low- from high-grade glioma is high, as illustrated inFig. 4.
FIG. 3. Classification accuracy (with SVM-RFE) vs number of re-tained features for classification of metastasis vs gliomas grade II,
Consistency in Feature Selection During Cross-Validation
III, or IV (1st row) and glioma grading (2nd row).
In order to assess if the same features are selected asdiscriminant features for each leave-one-out experiment,we calculated the normalized entropy (E) and standard
case of t test and N
deviation (S) of the ranking scores for each feature. Aver-
20 in the case of SVM-RFE). Figure
3 and Fig. 4 (1st row) show the classification accuracy with
aging of E and S across all selected features showed that
increasing number of retained features. The plots illustrate
the highest consistency was observed when distinguishing
that the fluctuations of accuracy around the optimal num-
meningioma from GBM (E ⫽ 0.95, S ⫽ 0.99, NF ⫽ 1), as
ber of features are small and the method is not very sen-
well as from gliomas grade II (E ⫽ 0.82, S ⫽ 0.92, NF ⫽ 9)
sitive to the exact number of selected features, except in
and metastasis (E ⫽ 0.71, S ⫽ 0.92, NF ⫽ 6).
the case of metastasis vs GBM, where the plateau is quitenarrow. The feature selection method in general elimi-
nates the redundant features, reduces the noise, and builds
For the multiclass problem, one-vs-all SVM classification
groupings that are both robust and accurate.
and majority voting are applied. For each one-vs-all clas-
Accuracy is relatively high for all classification pairs
sification task, feature selection is performed using SVM-
except for grade II vs grade III glioma. These two types of
RFE. Since in the multiclass problem the number of clas-
tumor have common characteristics due to the mixed his-
sifications to be performed is large, feature selection with
tology and are difficult to differentiate. On the other hand,
RFE on the total number of features (161) is computation-
the accuracy in classifying metastasis from primary glioma
ally very expensive. For this reason, we excluded from theevaluation all features that were never (or only once) se-lected as significant features in all binary classificationtasks (Table 2). Accordingly, 50 features were retained andused for feature selection. The confusion matrix withleave-one-out cross-validation is shown in Table 3. Menin-giomas were not included due to their small sample sizeand small clinical value. The highest classification accu-racy is achieved for metastasis, where only two (out of 24)samples are misclassified (one as glioma grade III and oneas GBM) and for low-grade tumors, with two (out of 22)cases being misclassified (one as glioma grade III and oneas metastasis).
DISCUSSION AND CONCLUSIONS
This paper presents a classification scheme for differenti-ating adult brain tumors using conventional MRI and rCBVmaps calculated from perfusion MRI. Shape characteris-tics, statistics on image intensities, and rotation-invariantGabor texture features are extracted from the central andmarginal tumoral, edematous, and necrotic region. Thescheme is fully automated and the help of an expert is not
FIG. 4. Classification accuracy (with SVM-RFE) vs number of re-
required, except for tracing the ROIs. Overall, we found
tained features (1st row) and receiver operating characteristic (ROC)
that SVM-based classification of texture patterns is a very
analysis (2nd row) for two main classification problems: metastasesvs primary gliomas (grades II, III, IV) shown in the 1st column and
promising approach to developing an objective and quan-
low- vs high-grade gliomas (grade II vs III and IV) shown in the 2nd
titative evaluation of brain tumors. However larger data-
sets need to be analyzed, which is expected to test the
Tumor Classification Using Machine Learning
generalization ability of this approach, but also to further
The feature subset selection method shows that all MR
improve its performance, as such classification systems
sequences are important for classification since different
perform better if trained more extensively.
features are selected for different classification tasks (Table
The results of multiclass classification (Table 3) illus-
2). The rCBV maps calculated from perfusion seem to be
trate that the highest classification accuracy is achieved for
particularly important since parameters extracted from
those are usually top ranked in most classification pairs.
whereas the classification accuracy for GBM is reduced
Also, contrast-enhanced T1 is a significant sequence; the
(29.4% are classified as grade III and 29.4% as metastasis).
volume of enhancement as percentage of total tumor vol-
The lowest classification rate in the multiclass problem is
ume (venh) is used as single feature for distinguishing be-
for the grade III glioma, where the largest portion (44.4%)
tween meningioma and GBM and is also part of the top-
is classified as grade II and the smaller portions as GBM
ranked features for other classification tasks. Texture cal-
(11.1%) or metastasis (11.1%). The prediction of glioma
culated on specific frequencies also contributes to tumor
grade is inherently difficult since brain neoplasms are
classification. Textural parameters extracted from the
often heterogeneous, meaning that different histopatho-
edematous area in FLAIR seem to be significant for glioma
logic features (such as mitotic index) can be present
grading, whereas texture in the neoplastic area in T1ce is
throughout an individual neoplasm. Therefore, part of our
important for distinguishing metastatic from glial tumors.
validation relies on examining the distribution of the false
The features currently used for classification are ex-
positives. The results of Table 3 show that most of the
tracted from the imaging profile and describe shape and
misclassified samples are assigned to a class of close de-
texture. We have not incorporated features describing the
gree of malignancy (grade). The degree of malignancy
deformation of the healthy structure due to tumor growth.
starts from the less malignant tumors (such as meningio-
It is known that different tumor types affect the surround-
mas) and increases until the high grade gliomas and me-
ing healthy region differently, and therefore studies have
tastases. The failure of the method to classify grade III
used the tumor mass effect as a descriptor for classifying
gliomas possibly indicates that the extracted features do
gliomas according to their clinical grade (20) or as an
not form a separate cluster, but are rather similar to the
independent predictor of survival (39). For this purpose,
features of the nearby classes (grade II and grade IV). The
we recently proposed an automated method for quantify-
binary classification tasks, where only tumors from two
ing the mass effect (40) by measuring how much the de-
single types are compared, exhibit higher classification
formation in the tumor origin deviates from the range
accuracy (mean ⫽ 91%; standard deviation ⫽ 7.7% over
observed in a normal population. We plan to incorporate
all tasks). The highest accuracy is achieved when distin-
the indicator of mass effect as an additional parameter in
guishing grade II glioma from metastasis (97.8%), and the
our classification framework in the future.
lowest, (75%) when distinguishing grade II from grade III
Moreover, a limitation of this framework comes from the
need for tracing ROIs, which makes the current approach
Generally, the classification accuracy using the pro-
semiautomatic and subject to intra- and interobserver vari-
posed method is comparable or higher than in other stud-
ability. However, it should be also noted that most of
ies (6) not using spectroscopy or diffusion tensor imaging.
applied features are based on spatial averaging and there-
The results in Devos et al. (21) using imaging intensities
fore are not very sensitive to small differences in the de-
from standard MR alone cannot be immediately compared
lineation of ROIs. The most sensitive features are expected
with ours because (i) they were based on ROIs extracted
to be the ones calculated over the marginal area of the
from spectroscopy imaging, which we want to avoid in
ROIs. We have investigated in the past the use of conven-
this study; and (ii) they reflected the number of voxels
tional and advanced MR imaging for automatic segmenta-
correctly classified rather than the number of subjects
tion of neoplastic and healthy tissue (41). We plan in the
(voxels from the same image were used as independent
future to combine the two frameworks in order to automat-
samples). In Georgiadis et al. (19), an SVM-based classifi-
ically segment and classify brain neoplasms. The aim of
cation system with radial basis function kernel achieved
this study is to assess the discrimination ability of stan-
74.4% overall accuracy in discriminating primary brain
dard MR imaging usually acquired in most clinical facili-
tumors from metastases utilizing the external cross-valida-
ties. The use of imaging intensities from standard MR
tion method, whereas an artificial neural network classifier
alone reaches lower performance than when combined
performed better (80% accuracy). The features employed
with spectroscopy (21). In the future, we plan to incorpo-
in that study were solely textural features from the T1ce
rate also diffusion tensor imaging and spectroscopy in our
MR images. Our analysis, achieving 84.7% accuracy on the
analysis for assessing the increase in classification accu-
leave-one-out cross-validation error, also showed that Ga-
racy. Other data based on microscopy imaging or his-
bor textural features from T1ce are important for this clas-
topathological examinations could also be included to in-
sification task combined with statistical parameters (mean,
crease accuracy in predictions. For example, nuclear fea-
variance, etc.) from other imaging sequences (T2, rCBV).
tures extracted from segmented nuclei (42) or blood vessel
Moreover, our results in glioma grading (low vs high
patterns (43) can assist brain astrocytoma malignancy
grade) are comparable with the ones reported in Li et al.
grading, and the spatial organization of tumor vessels can
(20) (the accuracy in Li et al. (20) was assessed by cross-
be indicative for differentiation between medulloblasto-
validation not being external to the feature-selection pro-
mas and supratentorial primitive neuroectodermal tumors
cess). This study was based on descriptive features esti-
(PNETs) (44). Voxel-wise histopathological parameters
mated by domain experts, whereas we applied features
that reflect proliferation and protein synthesis or PET im-
extracted in a semiautomatic way.
aging could also be used, if available, in order to extend
Zacharaki et al.
the current framework to output spatial maps of malig-
20. Li G, Yang J, Ye C, Geng D. Degree prediction of malignancy in brain
nancy (instead of a single grade), providing more reliable
glioma using support vector machines. Comput Biol Med 2006;36:313–
differentiation of heterogenous neoplasms.
21. Devos A, Simonetti AW, van der Graaf M, Lukas L, Suykens JA, Van-
hamme L, Buydens LM, Heerschap A, Van Huffel S. The use of multi-variate MR imaging intensities vs metabolic data from MR spectro-
scopic imaging for brain tumour classification. J Magn Reson 2005;173:
1. Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff
RM, Harsh GR, Cosgrove GR, Halpern EF, Hochberg FH, Rosen BR.
22. Tan TN. Rotation invariant texture features and their use in automatic
Cerebral blood volume maps of gliomas: comparison with tumor grade
script identification. IEEE Trans PAMI 1998;20:751–756.
and histological findings. Radiology 1994;191:41–51.
23. Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE,
2. Krabbe K, Gideon P, Wagn P, Hansen U, Thomsen C, Madsen F. MR
Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE,
diffusion imaging of human intracranial tumors. Neuroradiology 1997;
Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM,
39:483– 489.
Matthews PM. Advances in functional and structural MR image anal-
3. Provenzale JM, Mukundan S, Baroriak DP. Diffusion-weighted and
ysis and implementation as FSL. Neuroimage 2004;23(suppl 1):S208 –
perfusion MR imaging for brain tumor characterization and assessment
of treatment. Radiology 2006;239:632– 649.
24. Jenkinson M, Smith SA. Global optimisation method for robust affine
4. Wolde H, Pruim J, Mastik MF, Koudstaal J, Molenaar WM. Proliferative
registration of brain images. Med Image Anal 2001;5:143–156.
activity in human brain tumors: comparison of histopathology and
25. Smith SM: BET: brain extraction tool. FMRIB technical report
L-[l-HC] tyrosinePET. J Nucl Med 1997;38:1369 –1374.
5. Young RJ, Knopp EA. Brain MRI: tumor evaluation. J Magn Reson
26. Daugman JG. Uncertainty relations for resolution in space, spatial
Imaging 2006;24:709 –724.
frequency, and orientation optimized by two-dimensional visual corti-
6. Lev MH, Ozsunar Y, Henson JW, Rasheed AA, Barest GD, Harsh GR,
cal filters. J Opt Soc Am A 1985;A2:1160 –1169.
Fitzek MM, Chiocca EA, Rabinov JD, Csavoy AN, Rosen BR, Hochberg
27. Kruizinga P, Petkov N. Non-linear operator for oriented texture. IEEE
FH, Schaefer PW, Gonzalez RG. Glial tumor grading and outcome
Trans Image Process 1999;8:1395–1407.
prediction using dynamic spin-echo MR susceptibility mapping com-
28. Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects
pared with conventional contrast-enhanced MR: confounding effect of
of MRI acquisition parameter variations and protocol heterogeneity on
elevated rCBV of oligodendrogliomas. Am J Neuroradiol 2004;25:214 –
the results of texture analysis and pattern discrimination: an applica-
tion-oriented study. Med Phys 2009;36:1236 –1243.
7. Kremer S, Grand S, Remy C, Esteve F, Lefournier V, Pasquier B, Hoff-
29. Guyon I, Elisseeff A. An introduction to variable and feature selection.
mann D, Benabid AL, Le Bas JF. Cerebral blood volume mapping by MR
J Machine Learn Res 2003;3:1157–1182.
imaging in the initial evaluation of brain tumors. J Neuroradiol 2002;
30. Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical
recipes in C: the art of scientific computing. New York: Cambridge
8. Provenzale JM, Mukundan S, Baroriak DP. Diffusion-weighted and
University Press; 1992. 616 p.
perfusion MR imaging for brain tumor characterization and assessment
31. Breiman L. Bagging predictors. Machine Learn 1996;24:123–140.
of treatment response. Radiology 2006;239:632– 649.
32. Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer
9. Wang Q, Liacouras EK, Miranda E, Kanamala US, Megalooikonomou V.
classification using support vector machines. Machine Learn 2002;46:
Classification of brain tumors using MRI and MRS. In: Proceedings of
389 – 422.
the SPIE Conference on Medical Imaging, 2007.
33. Rakotomamonjy A. Variable selection using SVM-based criteria. J Ma-
10. Weber MA, Zoubaa S, Schlieter M, Ju¨ttler E, Huttner HB, Geletneky K,
chine Learn Res 2003;3:1357–1370.
Ittrich C, Lichy MP, Kroll A, Debus J, Giesel FL, Hartmann M, Essig M.
34. Zifeng C, Baowen X, Weifeng Z, Dawei J, Junling X. CLDA: feature
Diagnostic performance of spectroscopic and perfusion MRI for distinc-
selection for text categorization based on constrained LDA. Interna-
tion of brain tumors. Neurology 2006;66:1899 –1906.
tional Conference on Semantic Computing, p 702–712.
11. Cho Y-D, Choi G-H, Lee S-P, Kim J-K. 1H-MRS metabolic patterns for
35. Ambroise C, McLachlan GJ. Selection bias in gene extraction on the
distinguishing between meningiomas and other brain tumors. Magn
basis of microarray gene-expression data. Proc Natl Acad Sci USA
Reson Imaging 2003;21:663– 672.
12. Higano S, Yun X, Kumabe T, Watanabe M, Mugikura S, Umetsu A, Sato
36. Lachenbruch PA. Discriminant analysis. New York: Hafner Press; 1975.
A, Yamada T, Takahashi S. Malignant astrocytic tumors: clinical im-
37. Dasarathy BV, editor. Nearest neighbor (NN) norms: NN pattern clas-
portance of apparent diffusion coefficient in prediction of grade and
sification techniques. 1991.
prognosis. Radiology 2006;241:839 – 846.
38. Huang Y-M, Du S-X. Weighted support vector machine for classifica-
13. Al-Okaili RN, Krejza J, Woo JH, Wolf RL, O'Rourke DM, Judy KD,
tion with uneven training class sizes. Proceedings of 2005 International
Poptani H, Melhem ER. Intraaxial brain masses: MR imaging– based
Conference on Machine Learning and Cybernetics, 2005. p 4365– 4369.
diagnostic strategy—initial experience. Radiology 2007;243:539 –550.
39. Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F,
14. Tate AR, Majo´s C, Moreno A, Howe FA, Griffiths JR, Aru´s C. Automated
Lang FF, McCutcheon IE, Hassenbusch SJ, Holland E, Hess K, Michael
classification of short echo time in in vivo 1H brain tumor spectra: a
C, Miller D, Sawaya RA. Multivariate analysis of 416 patients with
multicenter study. Magn Reson Med 2003;49:29 –36.
glioblastoma multiforme: prognosis, extent of resection, and survival.
15. Majo´s C, Julia -Sape´ M, Alonso J, Serrallonga M, Aguilera C, Acebes JJ,
J Neurosurg 2001;95:190 –198.
Aru´s C, Gili J. Brain tumor classification by proton MR spectroscopy:
40. Zacharaki EI, Hogea CS, Shen D, Biros G, Davatzikos C. Non-diffeomor-
comparison of diagnostic accuracy at short and long TE. Am J Neuro-
phic registration of brain tumor images by simulating tissue loss and
radiol 2004;25:1696 –1704.
tumor growth. Neuroimage 2009;46:762–774.
16. Huang Y, Lisboa PJG, El-Deredy W. Tumour grading from magnetic
41. Verma R, Zacharaki EI, Ou Y, Cai H, Chawla S, Wolf R, Lee S-K,
resonance spectroscopy: a comparison of feature extraction with vari-
Melhem ER, Davatzikos C. Multi-parametric tissue characterization of
able selection. Stat Med 2003;22:147–164.
brain neoplasms and their recurrence using pattern classification of MR
17. Emblem KE, Zoellner FG, Tennoe B, Nedregaard B, Nome T, Due-
images. Acad Radiol 2008;15:966 –977.
Tonnessen P, Hald JK, Scheie D, Bjornerud A. Predictive modeling in
42. Glotsos D, Tohka J, Ravazoula P, Cavouras D, Nikiforidis G. Automated
glioma grading from MR perfusion images using support vector ma-
diagnosis of brain tumours astrocytomas using probabilistic neural
chines. Magn Reson Med 2008;60:945–952.
network clustering and support vector machines. Int J Neural Syst
18. Lu C, Devos A, Suykens JAK, Arus C, Van Huffel S. Bagging linear
sparse bayesian learning models for variable selection in cancer diag-
43. Selby DM, Woodard CA, Lakshman-Henry M, Bernstein JJ. Are endo-
nosis. IEEE Trans Informat Technol Med 2007;11:338 –346.
thelial cell patterns of astrocytomas indicative of grade? In Vivo 1997;
19. Georgiadis P, Cavouras D, Kalatzis I, Daskalakis A, Kagadis GC, Sifaki
K, Malamas M, Nikiforidis G, Solomou E. Improving brain tumor char-
44. Goldbrunner RH, Pietsch T, Vince GH, Bernstein JJ, Wagner S, Hage-
acterization on MRI by probabilistic neural networks and non-linear
man H, Selby DM, Krauss J, Soerensen N, Tonn JC. Different vascular
transformation of textural features. Comput Methods Programs Biomed
patterns of medulloblastoma and supratentorial primitive neuroecto-
2008;89:24 –32.
dermal tumors. Int J Dev Neurosci 1999;17:593–599.
Source: http://biosignal.med.upatras.gr/ezachar/papers/MRM_TumorClassif_2009.pdf
Basics of Molecular Cloning: Instructor's Manual Purpose and Concepts Covered .1 This instructor's ! manual is avaliable online only. This teaching resource is made available free ofcharge by Promega Corporation. Reproductionpermitted for noncommer- cial educational purposes only. Copyright 2010Promega Corporation. All rights reserved.
In Pharmacy, IMS MAT Jan 2011 To Fever and Pain INCLUDES DR. KEELY S TIPS Effective relief you can trust A Parent's Guide to Fever and Pain The content of this guide has been drafted in conjunction with Dr. JimKeely, who has spent 6 years working in Paediatrics at three of the mainteaching hospitals in Ireland. Dr Keely entered general practice in 1994and currently works as a GP at the Seabury Medical Centre in Malahidewith a special interest in Paediatrics. He is also the father of five childrenand gives us his personal top tips on how to deal with pain and fever.



