X96413u002
Cell, Vol. 103, 1121–1131, December 22, 2000, Copyright 2000 by Cell Press
Human Upf Proteins Target an mRNA for
Nonsense-Mediated Decay When Bound
Downstream of a Termination Codon
Jens Lykke-Andersen, Mei-Di Shu,
immunoglobulin and T cell receptor genes undergo so-
and Joan A. Steitz*
matic recombination and mutation to generate versatil-
Department of Molecular Biophysics
ity in the immune response. Truncated open reading
frames are abundant by-products, and premature termi-
Howard Hughes Medical Institute
nation codons in the T cell receptor- (TCR- ) gene lead
Yale University School of Medicine
to extraordinarily strong downregulation of the mRNA
295 Congress Avenue
(Carter et al., 1995, 1996; Li and Wilkinson, 1998).
New Haven, Connecticut 06536
The process of NMD has been well studied in yeast.
An mRNA containing a premature termination codon is
subjected to deadenylation-independent decapping by
Dcp1p, followed by 5⬘ to 3⬘ decay by the exonuclease
Xrn1p (Muhlrad and Parker, 1994; Beelman et al., 1996).
Nonsense-mediated decay (NMD) rids eukaryotic cells
A downstream sequence element (DSE) of low conser-
of aberrant mRNAs containing premature termination
vation, believed to appear throughout most coding re-
codons. These are discriminated from true termination
gions but never in the 3⬘ UTR, allows recognition of a
codons by downstream cis-elements, such as exon–
termination codon as premature (Peltz et al., 1993; Ha-
gan et al., 1995; Zhang et al., 1995). Recently, a heteroge-
exon junctions. We describe three novel human pro-
neous nuclear ribonucleoprotein (hnRNP), Hrp1p, was
teins involved in NMD, hUpf2, hUpf3a, and hUpf3b.
shown to interact with a DSE in vitro and may thus
While in HeLa cell extracts these proteins are com-
provide a downstream "mark" for a premature termina-
plexed with hUpf1, in intact cells hUpf3a and hUpf3b
tion codon (Gonza´lez et al., 2000).
are nucleocytoplasmic shuttling proteins, hUpf2 is
Nuclear as well as cytoplasmic events are involved
perinuclear, and hUpf1 cytoplasmic. hUpf3a and hUpf3b
in mammalian NMD. Not only are most mRNAs with
associate selectively with spliced  -globin mRNA in
premature termination codons less abundant in both com-
vivo, and tethering of any hUpf protein to the 3ⴕ UTR
partments than the corresponding wild-type mRNAs
of  -globin mRNA elicits NMD. These data suggest
(Cheng and Maquat, 1993; Belgrader and Maquat, 1994;
that assembly of a dynamic hUpf complex initiates in
Belgrader et al., 1994; Cheng et al., 1994; Kessler and
the nucleus at mRNA exon–exon junctions and triggers
Chasin, 1996; Zhang and Maquat, 1996; Zhang et al.,
NMD in the cytoplasm when recognized downstream
1998a), but pre-mRNA splicing is clearly involved. A
of a translation termination site.
termination codon is recognized as premature if located
upstream of an exon–exon junction (Cheng et al., 1994;
Carter et al., 1996; Thermann et al., 1998; Zhang et al.,
1998a, 1998b), with a distance of at least 50 nucleotides
Eukaryotic cells have evolved mechanisms to ensure
generally required (Cheng et al., 1994; Carter et al., 1996;
the fidelity of gene expression. One such mechanism,
Thermann et al., 1998; Zhang et al., 1998a, 1998b). Im-
called mRNA surveillance, makes certain that only
portantly, introns are rare in the 3⬘ UTR of mammalian
mRNAs with full coding potential are available for trans-
genes, and when they exist are nearly always within 50
lation in the cytoplasm (reviewed in Maquat, 1995; Ja-
nucleotides of the end of the open reading frame (Nagy
cobson and Peltz, 1996; Li and Wilkinson, 1998; Culbert-
and Maquat, 1998). Both general and mRNA-specific
son, 1999; Frischmeyer and Dietz, 1999; Hentze and
inhibition of translation blocks NMD (Belgrader et al.,
Kulozik, 1999; Hilleren and Parker, 1999a, 1999b). Aber-
1993; Carter et al., 1995; Thermann et al., 1998).
rant mRNAs containing premature termination codons
These observations raise the question of how a mam-
can arise from a failure in processing or by mutation.
malian cell determines that an intron, which was re-
They are recognized by the mRNA surveillance machin-
moved in the nucleus, was present downstream of a
ery and subjected to nonsense-mediated decay (NMD).
termination codon, which is later recognized by the ribo-
Thus, potentially deleterious truncated proteins are not
some and translation release factors in the cytoplasm.
expressed at high levels.
It has been hypothesized that splicing leaves a "mark"
Many human genetic disorders resulting from frame-
at or near exon–exon junctions that persists and acts
shift or nonsense mutations are believed to be rendered
post translation termination (Cheng et al., 1994; Maquat,
recessive by the NMD pathway. A well studied example
1995; Carter et al., 1996; Thermann et al., 1998; Zhang
is the human  -globin gene, where premature termina-
et al., 1998a; Hentze and Kulozik, 1999). Proteins that
tion codons in the first two exons are recessive, but
could constitute such a mark have recently been identi-
nonsense mutations in the third exon produce decay-
fied in HeLa nuclear extracts (Kataoka et al., 2000; Le
resistant mRNAs causing a dominant form of  -thalas-
Hir et al., 2000; McGarvey et al., 2000).
semia (Baserga and Benz, 1988; Thein et al., 1990;
Three extensively studied proteins in NMD are the
Enssle et al., 1993; Hall and Thein, 1994). mRNA surveil-
yeast polysome-associated Upf proteins: Upf1p, Upf2p/
lance is of particular importance to lymphocytes, where
Nmd2p, and Upf3p (Leeds et al., 1991, 1992; Atkin et
al., 1995, 1997; Cui et al., 1995; He and Jacobson, 1995;
* To whom correspondence should be addressed (e-mail: joan.
Lee and Culbertson, 1995). Upf1p is a cytoplasmic ATP-
dependent RNA helicase; mutations affecting its heli-
case activity also impair NMD (Czaplinski et al., 1995;
Weng et al., 1996a, 1996b, 1998). Upf1p interacts in two-
hybrid assays with Dcp2p/Nmd1p (He and Jacobson,
1995), a Dcp1p-interacting protein involved in cap re-
moval, suggesting a direct link between Upf1p and
mRNA decapping (Dunckley and Parker, 1999). In two-
hybrid assays, Upf2p interacts with both Upf1p and
Upf3p, and deletion of its Upf1p-interacting domain dis-
rupts NMD (He et al., 1996, 1997). Upf3p contains nu-
clear import and export signals and has been localized
to the nucleus (Shirley et al., 1998; Culbertson, 1999).
In addition to their role in NMD, yeast Upf proteins are
involved in translation termination. Their deletion en-
hances nonsense suppression (Leeds et al., 1992; Weng
et al., 1996a, 1996b, 1998; Maderazo et al., 2000), and
Upf1p interacts with translation release factors eRF1
and eRF3 (Czaplinski et al., 1998). In C. elegans seven
Smg genes are involved in NMD (Pulak and Anderson,
1993; Cali et al., 1999), with SMG-2 being a clear homo-
log of Upf1p (Page et al., 1999).
Here we identify and characterize human homologs
of yeast Upf2 and Upf3 proteins, hUpf2, hUpf3a, and
hUpf3b. Previously, a homolog of yeast Upf1p, hUpf1/
Rent1, had been reported to be involved in mammalian
NMD (Perlick et al., 1996; Applequist et al., 1997; Sun
et al., 1998), and very recently the human homolog of
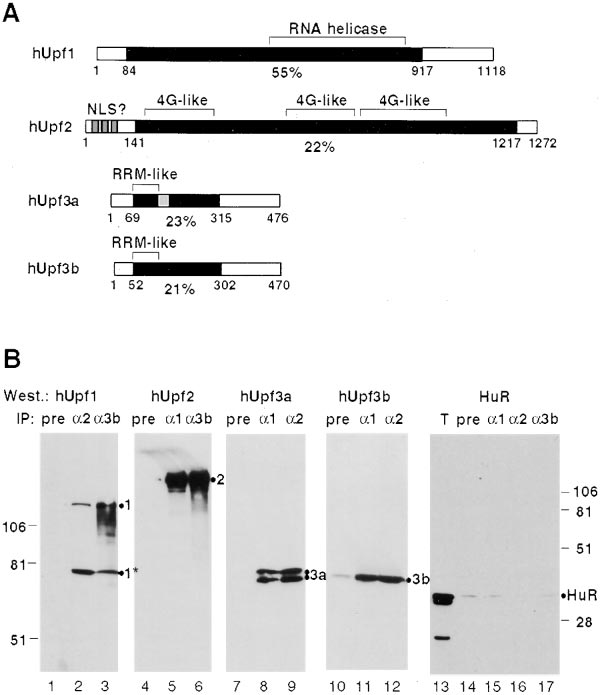
Figure 1. Human Upf Proteins Interact in HeLa Cell Extracts
Upf2 was identified and found to interact with hUpf1
(A) Schematic representation of human Upf proteins. Black boxes
represent regions of similarity to yeast Upf proteins, with the position
(Mendell et al., 2000). We demonstrate that binding of
within the protein (in amino acids) and the percent identity given
any hUpf protein downstream of a translation termina-
below. N and C termini, not similar to the corresponding yeast Upf
tion codon triggers NMD and that hUpf3a and hUpf3b
proteins, are shown as white boxes. The RNA helicase domain of
interact specifically with spliced mRNA in vivo. We find
hUpf1 is indicated. Putative nuclear localization signals (NLS) close
distinct subcellular locations for the human Upfs, sug-
to the N terminus of hUpf2 are depicted as gray boxes and eIF4G-
gesting that they do not simply form a complex that
like sequences, also present in yeast Upf2p (Ponting, 2000), are
indicated. The location of RRM-like sequences (20% identical to
scans the mRNA after translation termination. Rather, a
RRM-2 of ASF/SF2) in hUpf3a and hUpf3b are indicated and an
more dynamic surveillance complex begins assembly
alternative 99 nucleotide exon within hUpf3a is represented by a
on the mRNA in the nucleus and then directs NMD,
gray box. Sequences of hUpf2, hUpf3a, and hUpf3b are deposited in
probably at the perinuclear surface, when encountered
GenBank (accession numbers: AY013249, AY013250 and AY013251,
downstream of a site of translation termination.
respectively). Homologs of Upf2 and Upf3 can also be identified in
the completed genome sequences of Caenorhabditis elegans and
Drosophila melanogaster, as well as in Arabidopsis thaliana and
Schizosaccharomyces pombe (our unpublished observations). (B)
Coimmunoprecipitation of hUpf proteins from HeLa cell extracts
Identification of Human Homologs of Yeast Upf2
using anti-hUpf antibodies. hUpf proteins were immunoprecipitated
and Upf3 Proteins
from HeLa cell extracts using rabbit preimmune (pre; lanes 1, 4, 7,
To identify proteins involved in mammalian NMD, we
10 and 14), anti-hUpf1 (␣1; lanes 5, 8, 11 and 15), anti-hUpf2 (␣2;
lanes 2, 9, 12 and 16), or anti-hUpf3b antibodies (␣3b; lanes 3, 6
searched the nucleotide databases for human homologs
and 17). Immunoprecipitates, as well as one lane with 2.5% total
of yeast Upf2 and Upf3 proteins. The sequences of sev-
extract (T, lane 13), were electrophoresed on a 10% polyacrylamide/
eral expressed sequence tags (ESTs) were then used
SDS gel and, after transfer to a nitrocellulose membrane, probed
to design RT-PCR reactions, resulting in isolation of
with mouse polyclonal anti-hUpf1 (lanes 1–3), anti-hUpf2 (lanes 4–6),
cDNAs encoding proteins with limited similarity to yeast
anti-hUpf3a (lanes 7–9), anti-hUpf3b (lanes 10–12), or monoclonal
Upf2p and Upf3p. Human hUpf2, hUpf3a, and hUpf3b
anti-HuR antibodies (lanes 13–17). The hUpf1 (1), hUpf2 (2), hUpf3a
(3a), hUpf3b (3b), and HuR proteins are identified on the right of
are schematized in Figure 1A. hUpf2 contains several
each panel. Coelectrophoresed molecular weight markers are indi-
putative bipartite nuclear localization signals (NLS) close
cated on the left for lanes 1–12 and on the right for lanes 13–17. An
to its N terminus, as well as regions similar to eIF4G
ⵑ75 kDa degradation product of hUpf1, also present in the starting
also found in yeast Upf2p (Mendell et al., 2000; Ponting,
extract, is indicated by 1*. The two bands observed in lanes 8 and
2000). hUpf3a and hUpf3b contain a conserved domain
9 are most likely splice variants of hUpf3a that differ by 33 amino
with some similarity to an RNA recognition motif (RRM),
acids, as depicted in (A). hUpf3a and hUpf3b migrate unusually
slowly for their size (ⵑ52 kDa), probably due to a K/E-rich domain
indicating that they may be RNA binding proteins. The
(Graceffa et al., 1992).
RNP-2 motif within the RRM-like sequence is the most
conserved portion of all Upf3 proteins in the database
(our unpublished observations). Interestingly, the nu-
To demonstrate that hUpf proteins, like their yeast
clear export sequence identified in yeast Upf3p (Shirley
homologs, interact in cell extracts, rabbit and mouse
et al., 1998) overlaps with RNP-2, suggesting that nu-
polyclonal antibodies were raised against recombinant
clear export and RNA association could be linked.
hUpf1, hUpf2, hUpf3a, and hUpf3b. hUpf proteins immu-
Human Upf Proteins in Nonsense-Mediated Decay
1123
noprecipitated from freshly prepared RNase-treated
HeLa cell extracts using rabbit anti-hUpf antibodies
were subjected to Western blotting with mouse anti-
hUpf antibodies (Figure 1B). Relative to the amounts
present in the total extract (not shown) ⵑ2% of hUpf1
was coimmunoprecipitated with antibodies against
hUpf2 (lane 2), hUpf3a (not shown), or hUpf3b (lane 3),
while 20% of hUpf2, hUpf3a (both forms), and hUpf3b
coimmunoprecipitated with each of the other hUpf pro-
teins (lanes 5, 6, 8, 9, 11, 12). This is consistent with
Upf1p being about 10-fold more abundant than Upf2p
and Upf3p in yeast (Maderazo et al., 2000). Blotting with
a control monoclonal antibody against the RNA binding
protein HuR (Gallouzi et al., 2000) revealed that coimmu-
noprecipitation is specific (lanes 13–17). Antibodies
against hUpf3a and hUpf3b show low but significant
crossreaction, so it was not possible to test whether
these two proteins associate with each other.
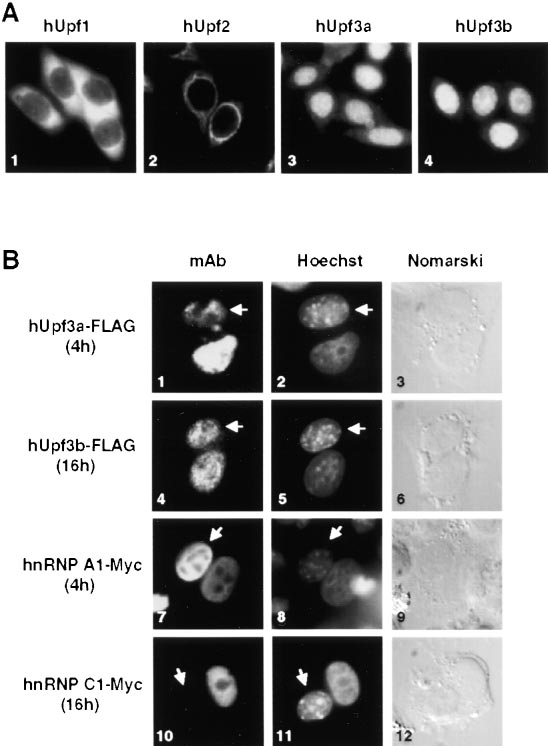
Human Upf1, Upf2, and Upf3 Proteins Are Concentrated
in Different Cellular Locations
The polyclonal antibodies raised against hUpf1, hUpf2,
hUpf3a, and hUpf3b were affinity purified and used to
localize endogenous hUpf proteins in HeLa cells. As
reported by Applequist et al. (1997), hUpf1 is evenly
distributed throughout the cytoplasm (Figure 2A, panel
1). In contrast, antibodies to hUpf2 show strong perinu-
Figure 2. Localization of hUpf Proteins in HeLa Cells
clear staining. hUpf3a and hUpf3b appear predomi-
(A) Immunocytochemical staining of fixed, permeabilized HeLa cells
nantly nuclear with a minor fraction in the cytoplasm
with affinity-purified rabbit anti-hUpf1 (panel 1), rabbit anti-hUpf2
(Figure 2A, panels 2–4).
(panel 2), mouse anti-hUpf3a (panel 3), or mouse anti-hUpf3b (panel
To test whether human Upf3 proteins shuttle in and
4) antibodies. Protein localizations were visualized using Texas Red
out of the nucleus, we performed heterokaryon experi-
conjugated anti-rabbit or Alexa 488 conjugated anti-mouse IgG anti-
ments (Pinol-Roma and Dreyfuss, 1992). HeLa cells tran-
bodies and confirmed using affinity purified antibodies from both
rabbit and mouse. (B) hUpf3a and hUpf3b are nucleocytoplasmic
siently transfected with plasmids expressing C-termi-
shuttling proteins. HeLa cells, transiently transfected with plasmids
nally FLAG-tagged hUpf3a or hUpf3b were fused with
producing FLAG-tagged hUpf3a (panels 1–3), FLAG-tagged hUpf3b
untransfected mouse L929 cells in the presence of the
(panels 4–6), Myc-tagged hnRNP A1 (panels 7–9), or Myc-tagged
translation inhibitor cycloheximide (Figure 2B). Four
hnRNP C1 (panels 10–12), were fused with mouse L929 cells using
hours after fusion, the hUpf3a-FLAG protein migrated
50% PEG-3350. After 4 hr (panels 1–3 and 7–9) or 16 hr (panels 4–6
partially from the HeLa cell nucleus to the mouse nu-
and 10–12) of incubation in the presence of cycloheximide, cells
were fixed and stained with monoclonal anti-FLAG (panels 1 and 4)
cleus (identified by spotted Hoechst staining and indi-
or anti-Myc (panels 7 and 10) antibodies. Protein localizations were
cated by an arrow, panels 1–3) and was equally distrib-
visualized using Alexa 488 conjugated anti-mouse IgG antibodies.
uted between the two nuclei by 16 hr after fusion (not
Human were distinguished from mouse nuclei (indicated by arrows)
shown). The hUpf3b-FLAG protein was not yet detect-
by staining with Hoechst dye 33258.
able in the mouse nucleus 4 hr after fusion (not shown),
but had fully equilibrated by 16 hr (panels 4–6). In control
experiments, Myc-tagged hnRNP A1, a shuttling protein,
the N or C terminus of an MS2 coat polypeptide defective
migrated to the mouse nucleus within 4 hr (panels 7–9),
in oligomerization (LeCuyer et al., 1995). Vectors ex-
whereas Myc-tagged hnRNP C1, a nonshuttling protein,
pressing these fusion proteins were cotransfected into
was absent from the mouse nucleus even after 16 hr
HeLa cells with constructs carrying the wild-type human
(panels 10–12). Shuttling of the hUpf3 proteins provides
-globin gene (with two introns), but containing in its
them with the potential to complex with the cytoplasmic
3⬘UTR two to six copies of an MS2 coat protein binding
hUpf1 and hUpf2 proteins.
site (starting 95 nucleotides downstream of the termina-
tion codon; Figure 3A, -2bs, -4bs and -6bs). Expres-
Human Upf3 Proteins Trigger mRNA Decay When
sion levels were controlled by inclusion of a third vector
Tethered to the 3ⴕUTR of -Globin mRNA
encoding -globin mRNA with an extended 3⬘UTR (with-
Since hUpf3a and hUpf3b are nucleocytoplasmic shut-
out MS2 coat protein binding sites; G). After 24 hr,
tling proteins and contain an RRM-like domain, they
cytoplasmic RNA was isolated and analyzed for -globin
could be part of a mark that targets an mRNA for NMD
mRNA by Northern blotting (Figure 3B).
when located downstream of a translation termination
The levels of -globin mRNAs with increasing num-
codon. We therefore tested whether tethering the hUpf3
bers of MS2 coat protein binding sites in their 3⬘UTRs
proteins to the 3⬘UTR of wild-type -globin mRNA (not
progressively decreased in cells coexpressing the hUpf3
containing a premature termination codon) would result
fusion proteins, up to 2-fold in the case of hUpf3a (Figure
in NMD. hUpf3a (long form) and hUpf3b were fused to
3B, lanes 2, 3, 7, 8, 12, and 13) and 5- to 6-fold with
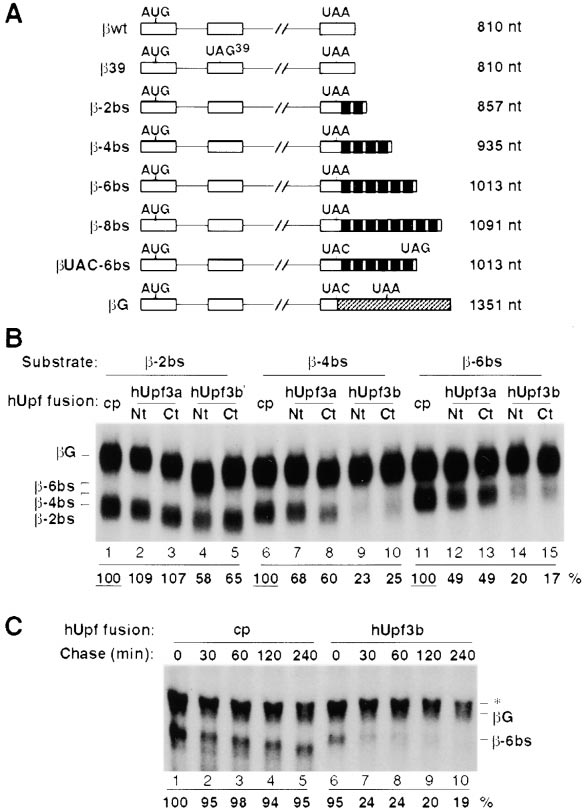
hUpf3b (Figure 3B, lanes 4, 5, 9, 10, 14, and 15). Coex-
pression of unfused MS2 coat protein (Figure 3B, lanes
1, 6, and 11) had no significant effect as compared to
the empty vector (see Figures 4C and 5B below). West-
ern blots using an anti-MS2 coat protein antibody (a
generous gift from Professor P. Stockley) revealed that
the hUpf3a fusion proteins are expressed at approxi-
mately 5- to 10-fold lower levels than the hUpf3b fusions
(data not shown), which could explain why the latter are
more active in the tethering assay. Immunocytochemical
experiments confirmed that, like their endogenous
counterparts (Figure 2), the MS2 coat protein fusions of
both hUpf3a and hUpf3b are nuclear and able to shuttle
(data not shown).
To test if the reduction in -globin mRNA levels trig-
gered by tethering hUpf3 proteins to the 3⬘UTR is due
to mRNA decay (as opposed to some other RNA proc-
essing event), a pulse–chase experiment was per-
formed. Plasmids containing the -globin gene with six
MS2 coat protein binding sites in the 3⬘UTR (-6bs) and
the control -globin gene (G), both under the control of
a tetracycline-responsive promoter, were cotransfected
with plasmids expressing either MS2 coat protein alone
or in N-terminal fusion with hUpf3b into HeLa "Tet-off"
cells in the presence of tetracycline (see Experimental
Procedures). hUpf3b was chosen for this experiment
because preliminary data showed that a strong effect
was required to obtain a clear result. Sixteen hours later,
a 4 hr transcriptional pulse was induced by washing
Figure 3. hUpf3a and hUpf3b Trigger NMD When Tethered to the
away the tetracycline. After addition of tetracycline to
3⬘UTR of -Globin mRNA
block further transcription, total -globin mRNA levels
(A) Schematic representation of -globin mRNAs expressed from
were monitored over time by Northern blotting.
plasmids used for transient transfection in Figures 3–5 (not to scale).
Exons and introns are shown as boxes and lines, respectively. Posi-
As seen in Figure 3C, tethering hUpf3b to the 3⬘UTR of
tions of translation initiation and termination codons are indicated
-globin mRNA leads to a biphasic mRNA decay pattern:
above each sequence. In UAC-6bs and G, the -globin termina-
ⵑ80% of the mRNA disappears within the first 30 min
tion codon (UAA) has been mutated to a UAC codon, as shown.
after blocking transcription (compare lanes 6 and 7),
Each MS2 coat protein binding site (19 nt) is depicted as a black
whereas the remaining ⵑ20% of the mRNA decays with
box. A 645 nt fragment of the GAPDH ORF and 3⬘UTR inserted
the same slow rate as the stable control mRNA (lanes
into G is cross-hatched. The lengths in nucleotides of the spliced
mRNAs (excluding their polyadenylate tails) are given on the right.
7–10). In contrast, -globin mRNA was stable when
(B) Tethering of hUpf3a and hUpf3b to the 3⬘UTR of -globin mRNA.
coexpressed with the MS2 coat protein (lanes 1–5).
HeLa cells were cotransfected with plasmids expressing -2bs
Gradual shortening of the -globin mRNA over time indi-
(lanes 1–5), -4bs (lanes 6–10), or -6bs (lanes 11–15) and G (lanes
cates that some deadenylation takes place (compare
1–15), as well as plasmids producing MS2 coat protein alone (cp,
-6bs in lanes 1–5). We conclude that tethering hUpf3b
lanes 1, 6, and 11) or fused to the N terminus (Nt) or C terminus (Ct)
to the 3⬘UTR of -globin mRNA induces rapid mRNA
of hUpf3a (lanes 2, 7, 12 and 3, 8, 13, respectively) or hUpf3b (lanes
4, 9, 14 and 5, 10, 15, respectively). Total HeLa cytoplasmic RNA
was prepared 24 hr after transfection, fractionated in a 1.2% formal-
dehyde/agarose gel, and probed with an antisense -globin cRNA
Tethering hUpf2 and hUpf1 to the 3ⴕUTR Also
after transfer to a nylon membrane. The identities of the different
Results in Decreased -Globin mRNA Levels
mRNAs are on the left. Band intensities were quantitated by phos-
Since the hUpf proteins interact in cell extracts (Figure
phorimager with the relative steady state level of the mRNA normal-
ized to the internal control (
1B), we asked whether tethering hUpf2 and hUpf1 to
G) and to the corresponding unfused
coat protein experiment, in which mRNA levels were set at 100%
-globin mRNA would also result in reduced mRNA lev-
(underlined). (C) A pulse–chase experiment demonstrates mRNA
els. Plasmids expressing these proteins fused to MS2
decay in the tethering assay. HeLa "Tet-off" cells were cotransfected
coat protein and -globin mRNAs with two to eight MS2
with plasmids expressing -6bs and G from a tetracycline-respon-
coat protein binding sites in their 3⬘UTRs were cotrans-
sive CMV promoter, as well as plasmids producing MS2 coat protein
fected. Similar to the hUpf3 proteins, tethering hUpf2
alone (cp, lanes 1–5) or fused to the N terminus of hUpf3b (lanes
6–10). After a 4 hr transcriptional pulse, cells were incubated for
(Figure 4A) or hUpf1 (Figure 4B) led to a 3-fold or 4-fold
various times as indicated above, and total RNA was subjected to
decreased level, respectively, of the reporter mRNA with
Northern blotting and quantitated as described above. The identities
eight binding sites. Western blots and immunocyto-
of the different mRNAs are on the right. A cross-hybridizing RNA
chemical staining revealed that the hUpf1 fusion protein
species (*) migrating just above G mRNA is seen in all experiments
was cytoplasmic and expressed at high levels similar
(e.g., see Figure 4A) but is especially apparent here, where the
to hUpf3b, whereas the hUpf2 fusion was even lower in
short transcriptional pulse results in much lower reporter RNA levels.
These RNAs were, however, sufficiently separated to allow accurate
abundance than hUpf3a (data not shown).
quantitation of the control G mRNA.
Mutant hUpf2 and hUpf1 proteins were also tested.
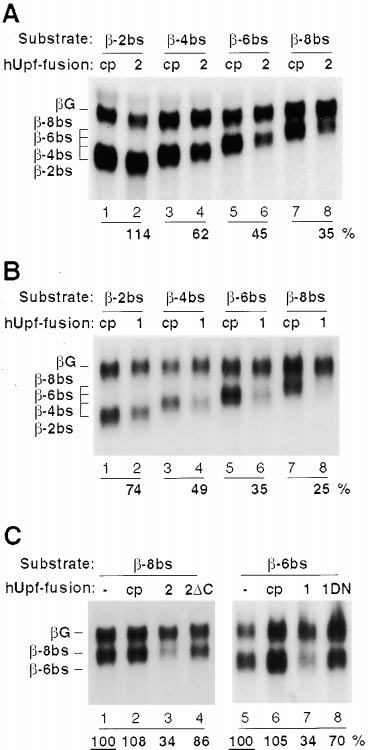
Human Upf Proteins in Nonsense-Mediated Decay
1125
when positioned within the 3⬘UTR of -globin mRNA.
Moreover, activity requires the putative hUpf1-inter-
acting domain of hUpf2, as well as an active helicase
domain in hUpf1.
Human Upf Protein-Triggered mRNA Reduction
Shows the Characteristics of NMD
We next asked if -globin mRNA reduction mediated by
tethering hUpf1, hUpf2, hUpf3a, or hUpf3b to the 3⬘UTR
downstream of the normal stop codon is in fact NMD.
Because NMD in mammalian cells requires active trans-
lation (Belgrader et al., 1993; Carter et al., 1995;
Thermann et al., 1998), the translation inhibitors cyclo-
heximide (Figure 5A) and pactamycin (not shown) were
tested. Cycloheximide had a general stabilizing effect
on all mRNA substrates in Figure 5A (compare – and ⫹
lanes), which was expected since most eukaryotic
mRNAs are somewhat stabilized when translation is in-
hibited (Herrick et al., 1990; Sachs, 1993). However, both
the -globin mRNA with a premature termination codon
(lanes 3, 4) and -globin reporters destabilized by teth-
ered hUpf proteins (lanes 7–14) were stabilized to a
greater extent than control mRNAs (lanes 1, 2, 5, 6).
Western blotting showed that this was not due to a
decrease in the levels of fusion proteins occurring during
the 2 hr incubation with translation inhibitors (data not
shown). Although cycloheximide-mediated stabilization
Figure 4. hUpf2 and hUpf1 Proteins Trigger NMD When Tethered
could be due to either lack of active translation or to
to the 3⬘UTR of -Globin mRNA
involvement of a labile protein factor, we conclude that
(A) Tethering of hUpf2 to the 3⬘UTR of -globin mRNA. HeLa cells
hUpf-mediated mRNA reduction behaves like NMD with
were cotransfected with plasmids expressing -2bs (lanes 1, 2),
respect to universal translation inhibition.
-4bs (lanes 3, 4), -6bs (lanes 5, 6), or -8bs (lanes 7, 8) and G
(lanes 1–8), as well as plasmids producing MS2 coat protein alone
We then investigated the importance of the position
(cp, lanes 1, 3, 5, and 7) or N-terminally fused to hUpf2 (2; lanes 2,
of the termination codon relative to the tethering sites
4, 6, and 8). Total HeLa cytoplasmic RNA was subjected to Northern
for the hUpf proteins. In -globin mRNA, insertion of an
blotting and quantitated as described in the legend to Figure 3B,
intron into the 3⬘UTR had been shown to lead to NMD
with mRNA identities indicated on the left. (B) Tethering of hUpf1
of an otherwise normal transcript, whereas mutation of
to the 3⬘UTR of -globin mRNA. Same experiment as in (A) but with
the termination codon (resulting in translation into the
hUpf1 fused to MS2 coat protein (1). (C) Mutant hUpf2 and hUpf1
proteins are impaired in triggering NMD. HeLa cells were cotrans-
3⬘UTR) stabilized the transcript (Thermann et al., 1998).
fected with plasmids expressing -8bs (lanes 1–4) or -6bs (lanes
We therefore produced a point mutation in the -globin
5–8) and G (lanes 1–8), as well as an empty pcDNA3 vector (-;
mRNA termination codon (UAA to UAC) that would result
lanes 1, 5) or a plasmid producing MS2 coat protein alone (cp, lanes
in translation termination downstream of the MS2 coat
2, 6) or coat protein N-terminally fused to hUpf2 (2; lane 3), to a
protein binding sites (see Figure 3A; UAC-6bs). When
hUpf2 deletion mutant lacking the C-terminal 204 amino acids (2⌬C;
cotransfected with MS2 coat protein fusions of hUpf3
lane 4), to hUpf1 (1; lane 7) or to a hUpf1 R844C (dominant-negative)
mutant (1DN; lane 8). Relative steady state levels of the
(Figure 5B), as well as hUpf2 and hUpf1 (not shown),
reporter mRNAs, calculated as above, are given below, with mRNA
this transcript was stable (lanes 7, 8, compare to 3,
identities indicated on the left.
4). Ribosomes most likely displace the tethered hUpf
proteins during translation, demonstrating that mRNA
reduction by hUpf tethering, similar to NMD, is a post-
Deletion of the C-terminal 206 amino acids of hUpf2, a
translational process. The transcript was not stabilized
region that in yeast Upf2p interacts with Upf1p (He et
simply by an MS2 coat protein–dependent blockage of
al., 1996; He et al., 1997), almost completely abolished
the translating ribosome, since insertion of an intron
hUpf2-triggered reduction in reporter mRNA levels (Fig-
downstream of the new termination codon destabilized
ure 4C, lane 4). Similarly, a point mutation in the helicase
the transcript both in the presence and absence of an
domain of hUpf1 (R844C, see below) impaired its effect
hUpf-MS2 fusion protein (data not shown).
(Figure 4C, lane 8). Western blotting revealed that the
Further evidence that hUpf1, hUpf2, and hUpf3 pro-
mutant fusion proteins were expressed at levels similar
teins trigger NMD was obtained using a dominant-nega-
to their wild-type counterparts (data not shown). In con-
tive hUpf1 mutant. Overexpression of this protein, which
trol experiments, a cytoplasmic RNA helicase, eIF4A,
contains a point mutation (R844C) in its RNA helicase
and two nuclear RNA binding proteins, hnRNP-A1 and
domain, was reported to yield a low (ⵑ2-fold) but repro-
hnRNP-C, had no effect when tethered to the 3⬘UTR of
ducible increase in the levels of mRNAs containing pre-
-globin mRNA (data not shown).
mature termination codons (Sun et al., 1998). Constructs
These results demonstrate that, like an intron, hUpf3,
expressing wild-type or dominant-negative hUpf1 were
hUpf2, or hUpf1 protein triggers reduced mRNA levels
cotransfected with plasmids expressing hUpf1, hUpf2,
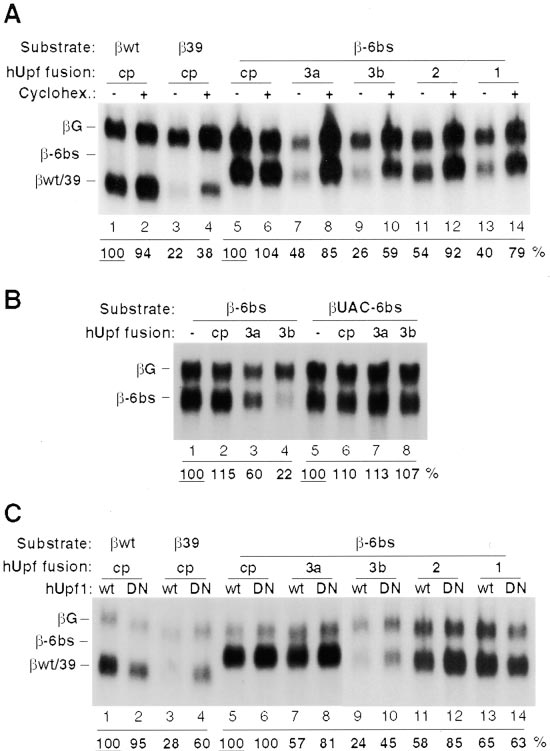
imental Procedures) resulted in less reduction in mRNA
levels in some cases (compare Figure 5C to Figures
3, 4, 5A, and 5B). Nevertheless, the dominant-negative
hUpf1 protein reproducibly enhanced (about 1.5- to
2-fold) the levels of the reporter -globin mRNA in the
presence of hUpf2- (lanes 11, 12) or hUpf3-fusions (lanes
7–10), whereas no effect was observed when the MS2
coat protein alone was tethered to the 3⬘UTR (lanes
5, 6). When hUpf1 instead was tethered downstream,
overexpression of the dominant-negative hUpf1 R844C
mutant protein had no effect (lanes 13, 14). Together
these results show that hUpf1, hUpf2, and hUpf3 pro-
teins are involved in NMD and that a termination codon
is recognized as premature when any of these proteins
binds downstream in the mRNA.
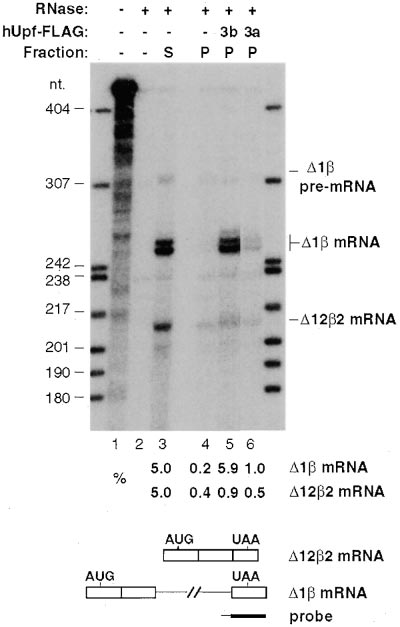
hUpf3a and hUpf3b Interact Specifically
with Spliced mRNA In Vivo
Our observation that hUpf proteins direct NMD when
tethered downstream of a translation termination codon
strongly suggests that these proteins normally interact
with exon–exon junctions in cellular mRNAs. To test if
hUpf3 proteins bind selectively to spliced mRNAs, two
constructs containing a CMV minimal promoter with tet-
racycline-responsive elements were made that express:
(1) intron-less -globin mRNA with a shortened 3⬘UTR
(⌬122) and (2) -globin mRNA containing intron 2, but
Figure 5. hUpf Protein-Triggered Decay Is Sensitive to Conditions
lacking intron 1 (⌬1) (Figure 6). They were cotrans-
fected with plasmids expressing FLAG-tagged hUpf3a
(A) hUpf protein-mediated -globin mRNA decay is inhibited by
or hUpf3b into HeLa "Tet-on" cells (see Experimental
cycloheximide. HeLa cells were cotransfected with plasmids ex-
Procedures) that contain a doxycycline-inducible tetra-
pressing wt (lanes 1, 2), 39 (lanes 3, 4) or -6bs (lanes 5–14) and
cycline transactivator. Twenty-four hours after transfec-
the G control (lanes 1–14), as well as an empty pcDNA3 vector (-;
lanes 1–4) or plasmids producing MS2 coat protein alone (cp, lanes
tion, doxycycline was added to induce -globin mRNA
5, 6) or coat protein N-terminally fused to hUpf3a (3a; lanes 7, 8),
expression, and cycloheximide was included to block
to hUpf3b (3b; lanes 9, 10), to hUpf2 (2; lanes 11, 12), or to hUpf1
translation as well as late steps in NMD that might lead
(1; lanes 13, 14). Twenty-four hours after transfection, cells were
to dissociation of the hUpf-mRNA complex. Cell extracts
incubated in the presence (⫹) or absence (-) of cycloheximide (50 g/
were prepared and RNAs immunoprecipitated by mono-
ml) for 2 hr. Total HeLa cytoplasmic RNA was subjected to Northern
clonal anti-FLAG antibodies were analyzed by RNase
blotting and quantitated as described in the legend to Figure 3B, with
mRNA identities indicated on the left. (B) Translation termination
protection for -globin mRNA.
downstream of the hUpf tethering sites abrogates -globin mRNA
The results in Figure 6 demonstrate that hUpf3b (lane
decay. HeLa cells were cotransfected with plasmids expressing
5) and hUpf3a (lane 6) interact selectively with -globin
-6bs (lanes 1–4) or UAC-6bs (lanes 5–8) and G (lanes 1–8) (see
mRNA that has undergone splicing. Approximately 6%
Figure 3A), as well as an empty pcDNA3 vector (-; lanes 1, 5) or
and 1% of spliced ⌬1 mRNA coprecipitated with
plasmids producing MS2 coat protein alone (cp; lanes 2, 6) or coat
hUpf3b and hUpf3a, respectively (compare lanes 5 and
protein N-terminally fused to hUpf3a (3a; lanes 3, 7) or to hUpf3b
(3b; lanes 4, 8). Total HeLa cytoplasmic RNA prepared 24 hr after
6 with 5% of the supernatant, lane 3). The lower activity
transfection was subjected to Northern blotting and quantitation as
of hUpf3a we believe is due to 5- to 10-fold lower expres-
described in the legend to Figure 3B, with mRNA identities indicated
sion levels of FLAG-tagged hUpf3a than FLAG-hUpf3b
on the left. (C) hUpf2, hUpf3a and hUpf3b, but not hUpf1 protein-
(data not shown). In contrast, after correction for back-
mediated -globin mRNA decay is sensitive to expression of a hUpf1
ground (lane 4), approximately 8- to 10-fold less of the
dominant-negative mutant. HeLa cells were cotransfected with plas-
intron-less ⌬122 mRNA was found in each of the anti-
mids expressing wt (lanes 1, 2), 39 (lanes 3, 4), or -6bs (lanes
5–14) and
hUpf3 immunoprecipitates (lanes 5, 6) and
G (lanes 1–14), as well as plasmids producing MS2 coat
protein alone (cp; lanes 1–6) or coat protein N-terminally fused to
mRNA was undetectable (a band deriving from the probe
hUpf3a (3a; lanes 7, 8), to hUpf3b (3b; lanes 9, 10), to hUpf2 (2;
alone, also present in lane 2, runs just below). Although
lanes 11, 12), or to hUpf1 (1; lanes 13, 14). A plasmid producing
the spliced and unspliced RNAs might differ in their rate
hUpf1 wild-type protein (wt; odd lanes) or dominant-negative hUpf1
of nuclear export, both are readily detectable in the
R844C mutant protein (DN; even lanes) was cotransfected in each
supernatant fraction (lane 3). We conclude that hUpf3a
case. Total HeLa cytoplasmic RNA prepared 24 hr after transfection
was subjected to Northern blotting and quantitation as described
and hUpf3b interact specifically with mRNAs containing
in the legend to Figure 3B.
exon–exon junctions in the absence of translation.
or hUpf3 fusion proteins and the -globin reporter con-
structs (Figure 5C). The lower amounts of hUpf fusion
We have characterized three novel human proteins in-
protein plasmids used in these transfections (see Exper-
volved in NMD, hUpf2, hUpf3a, and hUpf3b. They inter-
Human Upf Proteins in Nonsense-Mediated Decay
1127
(Figure 3C), (2) the hUpf-dependent reduction in reporter
mRNA levels is a posttranslational process because it
depends on active translation and because the tethering
site, like NMD cis-elements, must be downstream of the
termination codon (Figures 5A and 5B), and (3) overex-
pression of a dominant-negative hUpf1 mutant (Sun et
al., 1998) impairs the effects of tethering hUpf2 or hUpf3
(Figure 5C).
We believe that tethering the hUpf proteins down-
stream of a termination codon recapitulates a real step
in NMD, and that by fusing the hUpf proteins to the MS2
coat protein, coat protein binding sites become NMD
cis-elements. Since exon–exon junctions act as NMD
cis-elements, our data predict that hUpf proteins assem-
ble onto exon–exon junctions during, or after, pre-mRNA
splicing. Strongly supporting this interpretation is our
finding that hUpf3a and hUpf3b specifically interact with
a spliced -globin mRNA in vivo (Figure 6). In contrast
to other factors, like DEK and Y14, found deposited at
or near exon–exon junctions after mRNA splicing (Ka-
taoka et al., 2000; Le Hir et al., 2000; McGarvey et al.,
2000), we were not able to demonstrate a stable interac-
tion of hUpf3 proteins with adenovirus mRNA generated
Figure 6. hUpf3a and hUpf3b Specifically Interact with Spliced
in a HeLa splicing extract (data not shown). Perhaps
-Globin mRNA In Vivo
hUpf3 proteins join the multiprotein complex at exon–
HeLa "Tet-on" cells were cotransfected with plasmids expressing
exon junctions only at a very late step in mRNA matura-
⌬122 and ⌬1 RNAs (lanes 3–6; RNAs outlined below) and an
tion prior to export, which does not occur in vitro.
empty vector (-; lanes 3, 4) or a vector expressing FLAG-tagged
hUpf3b (3b; lane 5) or hUpf3a (3a; lane 6). 5% of total RNA from the
supernatant (S, lane 3) and 100% from the pellet (P; lanes 4–6)
A Perinuclear Surveillance Complex?
after anti-FLAG immunoprecipitation (see Experimental Procedures)
Coimmunoprecipitation experiments indicate that a sta-
were subjected to RNase protection assays for -globin mRNA.
ble complex containing hUpf1, hUpf2, and hUpf3 exists
RNase-treated samples were separated in a 5% polyacrylamide/
in RNase-treated HeLa cell extracts (Figure 1B). Surpris-
8M urea gel alongside control reactions of undigested (lane 1; 0.5%
loaded) and digested probe (lane 2; 100% loaded). The probe is
ingly, in intact cells each of the three hUpf proteins is
schematized below as a black bar (intron sequence thin). The identi-
concentrated in a different locale. hUpf3a and hUpf3b
ties of the different RNAs are indicated on the right. ⌬1-mRNA
are nucleocytoplasmic shuttling proteins, whereas hUpf2
routinely gives rise to a doublet in the RNase protection assay. Band
is perinuclear and hUpf1 is cytoplasmic, arguing that
intensities were quantitated by phosphorimager, and the relative
the hUpf complex most likely forms at the cytoplasmic
levels of the RNA species, given below, were normalized to the super-
surface of the nucleus. Perhaps the reason that hUpf3
natant fraction (lane 3), where 5% was loaded. Supernatants corre-
sponding to the pellet fractions in lanes 5 and 6 were indistinguish-
proteins require more than four hours to equilibrate fully
able from that shown in lane 3 (data not shown).
between the two nuclei in a heterokaryon [much longer
than other shuttling proteins such as hnRNP A1 and
HuR (Pinol-Roma and Dreyfuss, 1992; Fan and Steitz,
act in cell extracts with hUpf1, a previously identified
1998)] is that they normally never completely escape
protein in mammalian NMD (Perlick et al., 1996; Applequist
from the nuclear surface before they return.
et al., 1997; Sun et al., 1998; Mendell et al., 2000). We
The idea that the hUpf complex might form in the
observe that -globin reporter mRNAs are subjected to
perinuclear region helps resolve the controversy of
NMD when the hUpf proteins are tethered to their
whether NMD is a nuclear or cytoplasmic process (Ma-
3⬘UTRs via MS2 coat protein fusions, and that hUpf3
quat, 1995; Li and Wilkinson, 1998; Frischmeyer and
proteins interact selectively with spliced -globin mRNA
Dietz, 1999; Hentze and Kulozik, 1999). Most mRNAs
in vivo. The surprising finding that hUpf proteins exhibit
containing premature termination codons show de-
distinct subcellular locations suggests a model for the
creased abundance in the nuclear as well as the cyto-
assembly and action of the hUpf proteins on an mRNA
plasmic compartment (Cheng and Maquat, 1993; Bel-
targeted for NMD.
grader et al., 1994; Belgrader and Maquat, 1994; Cheng
et al., 1994; Kessler and Chasin, 1996; Zhang and Ma-
Human Upf Proteins Direct NMD When Bound
quat, 1996; Zhang et al., 1998a). These observations
Downstream of a Nonsense Codon
would be explained if aberrant mRNAs were degraded
We used tethering via the MS2 coat protein to demon-
while still attached to the nuclear surface, probably dur-
strate that human Upf proteins trigger NMD when bound
ing the first round of translation. Support for such local-
downstream of the termination codon of -globin mRNA.
ized degradation comes from the biphasic nature of the
Three criteria established that the lower levels of -glo-
decay seen in Figure 3C: the ⵑ20% of the reporter mRNA
bin mRNA observed upon hUpf tethering can be as-
that escapes initial rapid decay is as stable as wild-type
cribed to NMD: (1) pulse–chase experiments demon-
mRNA. This suggests that the mRNA may be checked
strated that tethering of hUpf3b results in mRNA decay
only once, during the first round of translation at the
the hUpf2/3p complex bound downstream results in
mRNA degradation (Figure 7). This explains why normal
mRNAs, which lack exon–exon junctions in their 3⬘UTRs
(and thus have all their hUpf3 displaced by translation
of their open reading frame), are not degraded by NMD.
Currently it is not clear whether hUpf1 joins the hUpf2/
3p complex before, during, or after translation termina-
tion; but active translation is required for hUpf1-medi-
ated decay in the tethering assay, so it is possible that
hUpf1 interacts with the hUpf2/3p complex before termi-
nation. How the hUpf complex then relays the signal for
selective decapping and decay of the aberrant mRNA
remains a problem for future investigation.
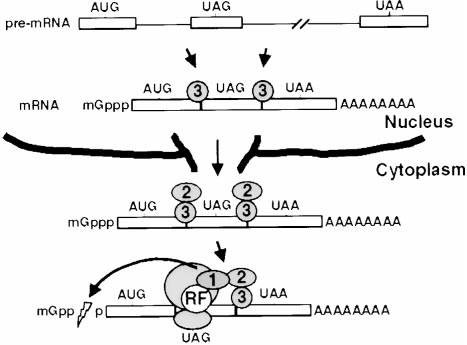
Figure 7. Model of the NMD Pathway
The order of events is described in the Discussion. An mRNA with
a premature termination codon in the second exon is schematized
with three boxes for exons and two lines for introns. hUpf1, hUpf2,
and hUp3 proteins are shown as circles with corresponding num-
Reporter plasmids used in this study all contain the human -globin
bers, and translation release factors are depicted by an encircled
gene, modified as schematized in Figure 3A (sequences available
"RF." A ribosome is shown as two elipses on top of each other.
upon request), and inserted between HindIII and XbaI sites of
Decapping of the mRNA is depicted by a lightning bolt in the cap
structure; mRNA degradation ensues.
Plasmids expressing N- and C-terminal MS2 coat protein fusions,
pcNMS2 and pcCMS2, contain an oligomerization-defective MS2
coat protein mutant from the construct pKCO-MS2CP-V75E;A81G
(LeCuyer et al., 1995), inserted into pcDNA3 with a modified polylin-
nuclear surface, and be either rapidly degraded or re-
ker. pcNMS2-hUpf1 and pcNMS2-hUpf1-R844C contain full-length
leased to the cytoplasm for further translation.
hUpf1 cDNA, the latter with a CGG to TGT mutation in codon 844.
pcNMS2-hUpf2 and pcNMS2-hUpf2⌬C contain hUpf2 cDNA ex-
tending from the translation initiation codon to the translation termi-
Mechanism of NMD
nation codon (codon 1273) or to codon 1066, respectively. pcNMS2-
In yeast, the three Upf proteins have been proposed
hUpf3a, pcCMS2-hUpf3a, pcNMS2-hUpf3b, and pcCMS2-hUpf3b
to form a surveillance complex that, after translation
contain full-length hUpf3a or hUpf3b cDNAs. Vectors for expression
termination, scans the mRNA for the presence of a
of unfused hUpf1 proteins, pcDNA-hUpf1 and pcDNA-hUpf1-
downstream DSE. Our data argue that the hUpf proteins
R844C, were prepared by moving BamHI/NotI inserts from pcNMS2-
hUpf1 and pcNMS2-hUpf1-R844C, respectively, into pcDNA3.
participate in a more dynamic process, whereby the
Vectors pcDNA-hUpf3a-FLAG and pcDNA-hUpf3b-FLAG contain
surveillance complex partially assembles on the mRNA
full-length hUpf3a and hUpf3b cDNAs with C-terminal FLAG tags
before translation. Specifically, we predict that hUpf3
inserted into pcDNA3.
first interacts with exon–exon junctions in the nucleus;
Plasmids for bacterial production of recombinant hUpf fusion pro-
second, hUpf2 interacts with hUpf3; and third, hUpf1
teins with N-terminal glutathione S-transferase (GST) or hexahisti-
joins the complex, probably via translation release fac-
dine (His6) tags contain hUpf cDNAs inserted between the BamHI and
NotI sites of, respectively, pGEX-4T1 (Pharmacia) and pET-His-HMK
tors. The following observations support this scenario:
(Jensen et al., 1997) with a modified polylinker. pGEX-4T1-hUpf1-
(1) A dominant–negative hUpf1 mutant impairs decay
SalFS and pET-His-hUpf1-SalFS contain full-length hUpf1 cDNAs,
when hUpf3 or hUpf2, but not hUpf1, is tethered down-
where a frame-shift mutation was created at codon 416 by removal
stream of the stop codon (Figure 5C). This argues both
of a SalI site. pGEX-4T1-hUpf2Ct and pET-His-hUpf2Ct contain the
that only one copy of hUpf1 contributes to an active
C-terminal 206 codons of hUpf2 cDNA (codons 1067 to 1272). pGEX-
hUpf complex and that hUpf1 acts downstream of hUpf3
4T1-hUpf3a, pET-His-hUpf3a, pGEX-4T1-hUpf3b, and pET-His-
hUpf3b contain full-length hUpf3a and hUpf3b cDNAs.
and hUpf2 in the NMD pathway. (2) hUpf3, hUpf2, and
hUpf1 are nuclear, perinuclear, and cytoplasmic in loca-
Production and Affinity Purification of Antibodies
tion, respectively (Figure 2A). (3) In yeast, Upf3p inter-
Rabbit and mouse polyclonal antibodies were raised against hUpf1
acts with Upf2p, which in turn interacts with Upf1p (He
(amino acids 1–416), hUpf2 (C-terminal 206 amino acids), hUpf3a
et al., 1997). (4) Polysome association of yeast Upf2p
(full-length) and hUpf3b (full-length) proteins fused to N-terminal
requires the presence of Upf3p but not of Upf1p,
GST tags. Antibodies were affinity purified by adsorption to N-termi-
whereas Upf1p and Upf3p are each found on polysomes
nally His6-tagged proteins, conjugated to CNBr-activated Sepharose
(Amersham/Pharmacia).
independent of any other Upf protein (Atkin et al., 1997).
We propose the model shown in Figure 7 for mamma-
lian NMD, similar to one proposed earlier for yeast (Cul-
HeLa spinner cells, washed twice in PBS (20 mM potassium phos-
bertson, 1999). During or after splicing, but while still
phate pH 7.4, 130 mM NaCl), were incubated at 1.5 ⫻ 108 cells/ml
nuclear, hUpf3 binds the nascent mRNA at or near exon–
in isotonic lysis buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1
exon junctions. This interaction could be either direct
mM EDTA, 0.1% Nonidet-P40, 1 mM phenylmethyl sulfonyl fluoride
or mediated by additional protein factors. Upon mRNA
(PMSF), 1 M aprotinin and 1 M leupeptin) containing 200 g/ml
RNase A, for 16 hr at 4⬚C. Cell debris was precipitated by centrifuga-
export, a perinuclear hUpf2 protein joins hUpf3. During
tion and 100 l of rabbit anti-hUpf antibody, or preimmune antibody,
the first round of translation, probably still at the nuclear
was added to 1 ml of the cleared lysate together with 1 mM of
surface, the progressing ribosome displaces any hUpf2
dithiothreitol and 1 mM PMSF. Cell extracts were incubated with
and hUpf3 proteins bound upstream of the termination
antibodies for 1 hr at 4⬚C, before addition of 100 g of protein A
codon. Upon translation termination, capture of hUpf1
sepharose (Pharmacia), preswollen in NET-2 (50 mM Tris-HCl pH
by release factors leading to concurrent recognition of
7.5, 150 mM NaCl, 0.05% Nonidet-P40), followed by incubation at
Human Upf Proteins in Nonsense-Mediated Decay
1129
4⬚C for four hours. Beads were then washed seven times with NET-2
the 3⬘ splice site of intron 2 to one nucleotide upstream of the -glo-
and resuspended in 100 l of SDS sample buffer (50 mM Tris-
bin polyadenylation signal.
HCl pH 6.8, 100 mM DTT, 2% sodium dodecylsulfate [SDS], 20%
glycerol, 0.2 mg/ml bromophenol blue). 10 l of each sample was
subjected to SDS/polyacrylamide gel electrophoresis followed by
Western blotting using mouse anti-hUpf antibodies or an anti-HuR
We thank Dagmar Dertinger and Olke Uhlenbeck for an MS2 coat
monoclonal antibody (Gallouzi et al., 2000). hUpf proteins were visu-
protein clone and Jenny Baker and Peter Stockley for polyclonal
alized by ECL (Pharmacia).
rabbit anti-MS2 coat protein antibodies. Constructive comments on
the manuscript were provided by Gia Voeltz, Kazio Tycowski, and
Miles Wilkinson, who also generously supplied cells. This research
was supported by National Institutes of Health grant CA 16038.
Immunocytochemical staining of HeLa cells and preparation of het-
J. L. was supported by fellowships from the Danish Natural Science
erokaryon HeLa/L929 cells was performed as described (Fan and
Research Council and the Alfred Benzon Foundation. J. A. S. is an
Steitz, 1998). Anti-FLAG M2 and anti-Myc monoclonal antibodies
investigator of the Howard Hughes Medical Institute.
(Sigma) were used at 10 g/ml and at a 1:200 dilution, respectively.
Affinity purified mouse and rabbit anti-hUpf antibodies were used
Received September 1, 2000; revised November 20, 2000.
in the range of 50–1000 ng/ml. Secondary antibodies were Alexa
488-conjugated anti-mouse IgG and Texas Red-conjugated anti-
rabbit IgG antibodies (Molecular Probes), used at a concentration
of 2–6 g/ml. Hoechst dye 33258 (Sigma) was included with the
Applequist, S.E., Selg, M., Raman, C., and Jack, H.M. (1997). Cloning
secondary antibodies at 1 g/ml to visualize the nuclei.
and characterization of HUPF1, a human homolog of the Saccharo-
myces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic
Transient Transfection Assays
Acids Res. 25, 814–821.
HeLa cells were grown in DMEM medium (Gibco-BRL) containing
Atkin, A.L., Altamura, N., Leeds, P., and Culbertson, M.R. (1995).
10% fetal bovine serum (FBS, Gibco-BRL) in 35 mm dishes and
The majority of yeast UPF1 co-localizes with polyribosomes in the
transiently transfected using Lipofectamin reagent (Gibco-BRL) ac-
cytoplasm. Mol. Biol. Cell 6, 611–625.
cording to the manufacturer's directions. For most experiments, 0.3
Atkin, A.L., Schenkman, L.R., Eastham, M., Dahlseid, J.N., Lelivelt,
g of each of the -globin reporter plasmids were cotransfected
M.J., and Culbertson, M.R. (1997). Relationship between yeast poly-
with 1.0 g of pcNMS2-hUpf1, -hUpf2, or -hUpf3 plasmid. In each
ribosomes and Upf proteins required for nonsense mRNA decay. J.
experiment in Figure 5C, 0.3 g of the -globin reporter plasmids
Biol. Chem. 272, 22163–22172.
were cotransfected with 0.6 g of one pcNMS2-hUpf plasmid and
1.5 g pcDNA-hUpf1 plasmid. 24 hr after transfection, growth me-
Baserga, S.J., and Benz, E.J. (1988). Nonsense mutations in the
dium was removed and the cells were washed once with PBS.
human -globin affect mRNA metabolism. Proc. Natl. Acad. Sci.
Cytoplasmic RNA was prepared by incubating the cells in 200 l
USA 85, 2056–2060.
gentle lysis buffer (10 mM HEPES-KOH pH 7.4, 10 mM NaCl, 3 mM
Beelman, C.A., Stevens, A., Caponigro, G., LeGrandeur, T.E., Hat-
MgCl2, 0.5% Nonidet P-40) for 1 min and transferring the resulting
field, L., Fortner, D.M., and Parker, R. (1996). An essential component
cytoplasmic extract to 1 ml of Trizol (Gibco-BRL) for RNA isolation
of the decapping enzyme required for normal rates of mRNA turn-
according to the manufacturer. One quarter (ⵑ10 g) of total cyto-
over. Nature 382, 642–646.
plasmic RNA was run per lane on a formaldehyde-agarose gel for
Belgrader, P., and Maquat, L.E. (1994). Nonsense but not missense
Northern blotting and probed with an internally 32P-labeled antisense
mutations can decrease the abundance of nuclear mRNA for the
-globin RNA covering the entire open reading frame.
mouse major urinary protein, while both types of mutations can
For pulse–chase experiments, HeLa "Tet-off" cells (Clontech)
facilitate exon skipping. Mol. Cell. Biol. 14, 6326–6336.
were transfected as above, except that the -globin reporter plas-
Belgrader, P., Cheng, J., and Maquat, L.E. (1993). Evidence to impli-
mids contained a tetracycline responsive CMV promoter. After
cate translation by ribosomes in the mechanism by which nonsense
transfection, cells were incubated in the presence of 50 ng/ml tetra-
codons reduce the nuclear level of human triosephosphate iso-
cycline for 16 hr. A transcriptional pulse was produced by incubating
merase mRNA. Proc. Natl. Acad. Sci. USA 90, 482–486.
cells for 4 hr in the absence of tetracycline; 1 g/ml tetracycline
was then added and total RNA was prepared using Trizol (Gibco-
Belgrader, P., Cheng, J., Zhou, X., Stephenson, L.S., and Maquat,
BRL) at different time points.
L.E. (1994). Mammalian nonsense codons can be cis effectors of
nuclear mRNA half-life. Mol. Cell. Biol. 14, 8219–8928.
Cali, B.M., Kuchma, S.L., Latham, J., and Anderson, P. (1999). smg-7
is required for mRNA surveillance in Caenorhabditis elegans. Genet-
HeLa "Tet-on" cells (Clontech) grown in 150 mm dishes were trans-
ics 151, 605–616.
fected with 7.5 g pc⌬1wt-TET, 10 g pc⌬122wt-TET and 5 g
pcDNA-hUpf3a-FLAG, pcDNA-hUpf3b-FLAG or pcDNA3 using Lipo-
Carter, M.S., Doskow, J., Morris, P., Li, S., Nhim, R.P., Sandstedt,
fectamin reagent (Gibco-BRL) according to the manufacturer.
S., and Wilkinson, M.F. (1995). A regulatory mechanism that detects
Twenty-four hours after transfection, doxycycline (Sigma) and cyclo-
premature nonsense codons in T-cell receptor transcripts in vivo is
heximide (Sigma) were added to 1 g/ml and 50 g/ml, respectively,
reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270,
and cells were incubated for 2 hr. Cells were washed and scraped
in PBS containing 50 g/ml cycloheximide, and after pelleting they
Carter, M.S., Li, S., and Wilkinson, M.F. (1996). A splicing-dependent
were lysed for 10 min on ice in 400 l hypotonic lysis buffer (10 mM
regulatory mechanism that detects translation signals. EMBO J. 15,
Tris-HCl pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton-X100, 1
mM PMSF, 1 M aprotinin, 1 M leupeptin and 0.1 units/l RNasin
Cheng, J., and Maquat, L.E. (1993). Nonsense codons can reduce
[Boehringer]). NaCl was added to 150 mM, and the cleared lysate
the abundance of nuclear mRNA without affecting the abundance
was incubated with 100 l of anti-FLAG-M2 agarose (Sigma) for 4
of pre-mRNA or the half-life of cytoplasmic mRNA. Mol. Cell. Biol.
hr at 4⬚C. Beads were washed eight times with 1 ml cold NET-2 and
FLAG-tagged proteins were eluted with 200 l NET-2 containing 1
Cheng, J., Belgrader, P., Zhou, X., and Maquat, L.E. (1994). Introns
mg/ml FLAG peptide (Sigma) at 4⬚C for 2 hr. Total RNA was prepared
are cis effectors of the nonsense-codon-mediated reduction in nu-
from the supernatant and eluted fractions using Trizol (Gibco-BRL),
clear mRNA abundance. Mol. Cell. Biol. 14, 6317–6325.
and contaminating plasmid was removed by digestion with RQ1 DNase
(Promega). RNase protections were performed by standard proce-
Cui, Y., Hagan, K.W., Zhang, S., and Peltz, S.W. (1995). Identification
dures (Fan and Steitz, 1998), except that 2% polyvinyl alcohol was
and characterization of genes that are required for the accelerated
included during hybridization. The probe was an internally [32P]-labeled
degradation of mRNAs containing a premature translational termina-
antisense -globin RNA extending from 60 nucleotides upstream of
tion codon. Genes Dev. 9, 423–436.
Culbertson, M.R. (1999). RNA surveillance. Unforeseen conse-
Wang, F., and Dreyfuss, G. (2000). Pre-mRNA splicing imprints
quences for gene expression, inherited genetic disorders and can-
mRNA in the nucleus with a novel RNA-binding protein that persists
cer. Trends Genet. 15, 74–80.
in the cytoplasm. Mol. Cell 6, 673–682.
Czaplinski, K., Weng, Y., Hagan, K.W., and Peltz, S.W. (1995). Purifi-
Kessler, O., and Chasin, L.A. (1996). Effects of nonsense mutations
cation and characterization of the Upf1 protein: a factor involved in
on nuclear and cytoplasmic adenine phosphoribosyltransferase
translation and mRNA degradation. RNA 1, 610–623.
RNA. Mol. Cell. Biol. 16, 4426–4435.
Czaplinski, K., Ruiz-Echevarria, M.J., Paushkin, S.V., Han, X., Weng,
LeCuyer, K.A., Behlen, L.S., and Uhlenbeck, O.C. (1995). Mutants
Y., Perlick, H.A., Dietz, H.C., Ter-Avanesyan, M.D., and Peltz, S.W.
of the bacteriophage MS2 coat protein that alter its cooperative
(1998). The surveillance complex interacts with the translation re-
binding to RNA. Biochemistry 34, 10600–10606.
lease factors to enhance termination and degrade aberrant mRNAs.
Lee, B.S., and Culbertson, M.R. (1995). Identification of an additional
Genes Dev. 12, 1665–1677.
gene required for eukaryotic nonsense mRNA turnover. Proc. Natl.
Dunckley, T., and Parker, R. (1999). The DCP2 protein is required
Acad. Sci. USA 92, 10354–10358.
for mRNA decapping in Saccharomyces cerevisiae and contains a
Leeds, P., Peltz, S.W., Jacobson, A., and Culbertson, M.R. (1991).
functional MutT motif. EMBO J. 18, 5411–5422.
The product of the yeast UPF1 gene is required for rapid turnover
Enssle, J., Kugler, W., Hentze, M.W., and Kulozik, A.E. (1993). Deter-
of mRNAs containing a premature translational termination codon.
mination of mRNA fate by different RNA polymerase II promoters.
Genes Dev. 5, 2303–2314.
Proc. Natl. Acad. Sci. USA 90, 10091–10095.
Leeds, P., Wood, J.M., Lee, B.-S., and Culbertson, M.R. (1992). Gene
Fan, X.C., and Steitz, J.A. (1998). Overexpression of HuR, a nuclear-
products that promote mRNA turnover in Saccharomyces cerevis-
cytoplasmic shuttling protein, increases the in vivo stability of ARE-
iae. Mol. Cell. Biol. 12, 2165–2177.
containing mRNAs. EMBO J. 17, 3448–3460.
Le Hir, H., Moore, M.J., and Maquat, L.E. (2000). Pre-mRNA splicing
Frischmeyer, P.A., and Dietz, H.C. (1999). Nonsense-mediated
alters mRNP composition: evidence for stable association of pro-
mRNA decay in health and disease. Hum. Mol. Genet. 8, 1893–1900.
teins at exon-exon junctions. Genes Dev. 14, 1098–1108.
Gallouzi, I., Brennan, C.M., Stenberg, M.G., Swanson, M.S., Ever-
Li, S., and Wilkinson, M.F. (1998). Nonsense surveillance in lympho-
sole, A., Maizels, N., and Steitz, J.A. (2000). HuR binding to cyto-
cytes? Immunity 8, 135–141.
plasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci.
Maderazo, A.B., He, F., Mangus, D.A., and Jacobson, A. (2000).
USA 97, 3073–3078.
Upf1p control of nonsense mRNA translation is regulated by Nmd2p
Gonza´lez, C.I., Ruiz-Echevarri´a, M.J., Vasudevan, S., Henry, M.F.,
and Upf3p. Mol. Cell. Biol. 20, 4591–4603.
and Peltz, S.W. (2000). The yeast hnRNP-like protein Hrp1/Nab4
Maquat, L.E. (1995). When cells stop making sense: effects of non-
marks a transcript for nonsense-mediated mRNA decay. Mol. Cell
sense codons on RNA metabolism in vertebrate cells. RNA 1,
Graceffa, P., Jancso, A., and Mabuchi, K. (1992). Modification of
McGarvey, T., Rosonina, E., McCracken, S., Li, Q., Arnaout, R., Mien-
acidic residues normalizes sodium dodecyl sulfate-polyacrylamide
tjes, E., Nickerson, J.A., Awrey, D., Greenblatt, J., Grosveld, G.,
gel electrophoresis of caldesmon and other proteins that migrate
and Blencowe, B.J. (2000). The acute myeloid leukemia-associated
anomalously. Arch. Biochem. Biophys. 297, 46–51.
protein, DEK, forms a splicing-dependent interaction with exon-
Hagan, K.W., Ruiz-Echevarria, M.J., Quan, Y., and Peltz, S.W. (1995).
product complexes. J. Cell Biol. 150, 309–320.
Characterization of cis-acting sequences and decay intermediates
Mendell, J.T., Medghalchi, S.M., Lake, R.G., Noensie, E.N., and
involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 15,
Dietz, H.C. (2000). Novel Upf2p orthologues suggest a functional link
between translation initiation and nonsense surveillance complexes.
Hall, G.W., and Thein, S.L. (1994). Nonsense codon mutations in the
Mol. Cell. Biol. 20, 8944–8957.
terminal exon of the -globin gene are not associated with a reduc-
Muhlrad, D., and Parker, R. (1994). Premature translational termina-
tion in -mRNA accumulation: a mechanism for phenotype of domi-
tion triggers mRNA decapping. Nature 370, 578–581.
nant -thalassemia. Blood 83, 2031–2037.
Nagy, E., and Maquat, L.E. (1998). A rule for termination-codon
He, F., and Jacobson, A. (1995). Identification of a novel component
position within intron-containing genes: when nonsense affects RNA
of the nonsense-mediated mRNA decay pathway by use of an inter-
abundance. Trends Biochem. Sci. 23, 198–199.
acting protein screen. Genes Dev. 9, 437–454.
Page, M.F., Carr, B., Anders, K.R., Grimson, A., and Anderson, P.
He, F., Brown, A.H., and Jacobson, A. (1996). Interaction between
(1999). SMG-2 is a phosphorylated protein required for mRNA sur-
Nmd2p and Upf1p is required for activity but not for dominant-
veillance in Caenorhabditis elegans and related to Upf1p of yeast.
negative inhibition of the nonsense-mediated mRNA decay pathway
Mol. Cell. Biol. 19, 5943–5951.
in yeast. RNA 2, 153–170.
He, F., Brown, A.H., and Jacobson, A. (1997). Upf1p, Nmd2p, and
Peltz, S.W., Brown, A.H., and Jacobson, A. (1993). mRNA destabili-
Upf3p are interacting components of the yeast nonsense-mediated
zation triggered by premature translational termination depends on
mRNA decay pathway. Mol. Cell. Biol. 17, 1580–1594.
at least three cis-acting sequence elements and one trans-acting
factor. Genes Dev. 7, 1737–1754.
Hentze, M.W., and Kulozik, A.E. (1999). A perfect message: RNA
surveillance and nonsense-mediated decay. Cell 96, 307–310.
Perlick, H.A., Medghalchi, S.M., Spencer, F.A., Kendzior, R.J., Jr.,
and Dietz, H.C. (1996). Mammalian orthologues of a yeast regulator
Herrick, D., Parker, R., and Jacobson, A. (1990). Identification and
of nonsense transcript stability. Proc. Natl. Acad. Sci. USA 93,
comparison of stable and unstable mRNAs in Saccharomyces cere-
visiae. Mol. Cell. Biol. 10, 2269–2284.
Pinol-Roma, S., and Dreyfuss, G. (1992). Shuttling of pre-mRNA
Hilleren, P., and Parker, R. (1999a). mRNA surveillance in eukaryotes:
binding proteins between nucleus and cytoplasm. Nature 355,
kinetic proofreading of proper translation termination as assessed
by mRNP domain organization? RNA 5, 711–719.
Ponting, C.P. (2000). Novel eIF4G domain homologues linking mRNA
Hilleren, P., and Parker, R. (1999b). Mechanisms of mRNA surveil-
translation with nonsense-mediated mRNA decay. Trends Biochem.
lance in eukaryotes. Annu. Rev. Genet. 33, 229–260.
Sci. 25, 423–426.
Jacobson, A., and Peltz, S.W. (1996). Interrelationships of the path-
ways of mRNA decay and translation in eukaryotic cells. Annu. Rev.
Pulak, R., and Anderson, P. (1993). mRNA surveillance by the Caeno-
Biochem. 65, 693–739.
rhabditis elegans smg genes. Genes Dev. 7, 1885–1897.
Jensen, T.H., Jensen, A., Szilvay, A.M., and Kjems, J. (1997). Probing
Sachs, A.B. (1993). Messenger RNA degradation in eukaryotes. Cell
the structure of HIV-1 Rev by protein footprinting of multiple mono-
clonal antibody binding sites. FEBS Lett. 414, 50–54.
Shirley, R.L., Lelivelt, M.J., Schenkman, L.R., Dahlseid, J.N., and
Kataoka, N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A.,
Culbertson, M.R. (1998). A factor required for nonsense-mediated
Human Upf Proteins in Nonsense-Mediated Decay
1131
mRNA decay in yeast is exported from the nucleus to the cytoplasm
by a nuclear export signal sequence. J. Cell Sci. 111, 3129–3143.
Sun, X., Perlick, H.A., Dietz, H.C., and Maquat, L.E. (1998). A mutated
human homologue to yeast Upf1 protein has a dominant-negative
effect on the decay of nonsense-containing mRNAs in mammalian
cells. Proc. Natl. Acad. Sci. USA 95, 10009–10014.
Thein, S.L., Hesketh, C., Taylor, P., Temperley, I.J., Hutchinson,
R.M., Old, J.M., Wood, W.G., Clegg, J.B., and Weatherall, D.J. (1990).
Molecular basis for dominantly inherited inclusion body -thalas-
semia. Proc. Natl. Acad. Sci. USA 87, 3924–3928.
Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hage-
meier, C., Hentze, M.W., and Kulozik, A.E. (1998). Binary specifica-
tion of nonsense codons by splicing and cytoplasmic translation.
EMBO J. 17, 3484–3494.
Weng, Y., Czaplinski, K., and Peltz, S.W. (1996a). Identification and
characterization of mutations in the UPF1 gene that affect nonsense
suppression and the formation of the Upf protein complex but not
mRNA turnover. Mol. Cell. Biol. 16, 5491–5506.
Weng, Y., Czaplinski, K., and Peltz, S.W. (1996b). Genetic and bio-
chemical characterization of mutations in the ATPase and helicase
regions of the Upf1 protein. Mol. Cell. Biol. 16, 5477–5490.
Weng, Y., Czaplinski, K., and Peltz, S.W. (1998). ATP is a cofactor
of the Upf1 protein that modulates its translation termination and
RNA binding activities. RNA 4, 205–214.
Zhang, J., and Maquat, L.E. (1996). Evidence that the decay of
nucleus-associated nonsense mRNA for human triosephosphate
isomerase involves nonsense codon recognition after splicing. RNA
2, 235–243.
Zhang, S., Ruiz-Echevarria, M.J., Quan, Y., and Peltz, S.W. (1995).
Identification and characterization of a sequence motif involved in
nonsense-mediated mRNA decay. Mol. Cell. Biol. 15, 2231–2244.
Zhang, J., Sun, X., Qian, Y., LaDuca, J.P., and Maquat, L.E. (1998a).
At least one intron is required for the nonsense-mediated decay of
triosephosphate isomerase mRNA: a possible link between nuclear
splicing and cytoplasmic translation. Mol. Cell. Biol. 18, 5272–5283.
Zhang, J., Sun, X., Qian, Y., and Maquat, L.E. (1998b). Intron function
in the nonsense-mediated decay of beta-globin mRNA: indications
that pre-mRNA splicing in the nucleus can influence mRNA transla-
tion in the cytoplasm. RNA 4, 801–815.
GenBank Accession Numbers
Sequences of hUpf2, hUpf3a, and hUpf3b have been deposited
in GenBank under accession numbers AY013249, AY013250, and
AY013251, respectively.
Source: http://faculty.kfupm.edu.sa/CHEM/thukair/Reading/RA2/Human%20Upf%20Proteins%20Target%20an%20mRNA.pdf
Wiri Wai Care Wonders! Students from Wiri Central School proudly sign W after completing work on the Puhinui stream. Learning goes beyond the classroom Drury School drain painting Wai Care Fieldtrip to Hunua Brendon with a longfin eel found during fish monitoring Drury School's Wai Care club recently painted up a Papakura Normal School's middle and senior American Journal of Epidemiology ª The Author 2011. Published by Oxford University Press on behalf of the Johns Hopkins Bloomberg School of DOI: 10.1093/aje/kwq513 Public Health. All rights reserved. For permissions, please e-mail: [email protected]. Advance Access publication: Original Contribution Long-Term Sedentary Work and the Risk of Subsite-specific Colorectal Cancer
|
|








