Doi:10.1016/j.cell.2008.06.050
SIRT1 Regulates Circadian Clock GeneExpression through PER2 Deacetylation
Gad Asher,David Gatfield,Markus Hans Reinke,Charna Florian Kreppel,Raul Mostoslavsky,Frederick W. Alt,and Ueli 1Department of Molecular Biology, Sciences III, University of Geneva, 30, Quai Ernest Ansermet, CH-1211 Geneva-4, Switzerland2Division of Gene Therapy, University of Ulm, Helmholtzstrasse 8/1, D-89081 Ulm, Germany3Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA 02114, USA4Howard Hughes Medical Institute, Children's Hospital, Center for Blood Research, and Department of Genetics,Harvard University Medical School, Boston, MA 02115, USA*Correspondence: DOI 10.1016/j.cell.2008.06.050
The molecular oscillator in both master and subsidiary clocks
(is thought to rely on
The mammalian circadian timing system is com-
a negative transcriptional feedback loop
posed of a central pacemaker in the suprachiasmatic
nucleus of the brain that synchronizes countless
PAS domain helix-loop-helix proteins BMAL1 and CLOCK (or
subsidiary oscillators in peripheral tissues. The
its paralog NPAS2 [])
rhythm-generating mechanism is thought to rely on
bind as heterodimers to regulatory elements of
Cry and
Per genesand stimulate the transcription of these genes. Once the repres-
a feedback loop involving positively and negatively
sor proteins CRY and PER have reached a critical concentration,
acting transcription factors. BMAL1 and CLOCK
they attenuate the activity of BMAL1-CLOCK heterodimers and
activate the expression of Period (Per) and Crypto-
thereby repress the transcription of their own genes. In addition,
chrome (Cry) genes, and once PER and CRY proteins
an interconnecting feedback loop involving orphan nuclear re-
accumulate to a critical level they form complexes
ceptors of the REV-ERB and ROR families regulates the expres-
with BMAL1-CLOCK heterodimers and thereby
sion of
Bmal1 ).
repress the transcription of their own genes. Here,
Several lines of evidence suggest a strong interplay between
we show that SIRT1, an NAD+-dependent protein
metabolism and the circadian clock (
deacetylase, is required for high-magnitude circa-
dian transcription of several core clock genes, in-
feeding cycles as a
Zeitgeber for peripheral clocks implies that
cluding Bmal1, Rorg, Per2, and Cry1. SIRT1 binds
the circadian clock plays an important role in nutrient process-ing and energy homeostasis. Indeed, transcriptome profiling
CLOCK-BMAL1 in a circadian manner and promotes
studies revealed that many genes involved in metabolism
the deacetylation and degradation of PER2. Given
are rhythmically expressed (
the NAD+ dependence of SIRT1 deacetylase activity,
it is likely that SIRT1 connects cellular metabolism to
Furthermore, at least
the circadian core clockwork circuitry.
in vitro, the DNA-binding activity of BMAL1-CLOCK is stronglyinfluenced by the ratio of reduced to oxidized NAD cofactors,
which are often considered as a readout of the cellular metabolicstate ).
The physiology and behavior of mammals are subject to daily
SIRT1 is the mammalian homolog of yeast Sir2, an NAD+-
oscillations driven by an endogenous circadian clock (
dependent deacetylase involved in transcriptional silencing,
genome stability, and longevity (
the circadian timing system is composed of a central pacemaker
). The SIRT1 catalytic reaction involves
in the brain's suprachiasmatic nucleus (SCN) and subsidiary
the breakdown of one NAD+ molecule for each deacetylated
oscillators in most peripheral tissues. While light-dark cycles
acetyl lysine and the generation of nicotinamide and O-acetyl-
are the predominant
Zeitgebers (timing cues) for the SCN pace-
ADP-ribose. SIRT1 was found to deacetylate not only histones
maker, cyclic feeding behavior is a strong
Zeitgeber for clocks
but also several transcriptional regulatory proteins involved in
operating in many peripheral tissues
the control of metabolism, including members of the FOXO
). It is therefore likely that the SCN synchronizes
peroxisome proliferator-activated receptor gamma
peripheral oscillators by imposing rest-activity rhythms and
(PPARg) coactivator 1a (PGC1a), and the nuclear receptor LXR
thus feeding-fasting cycles.
Cell
134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 317
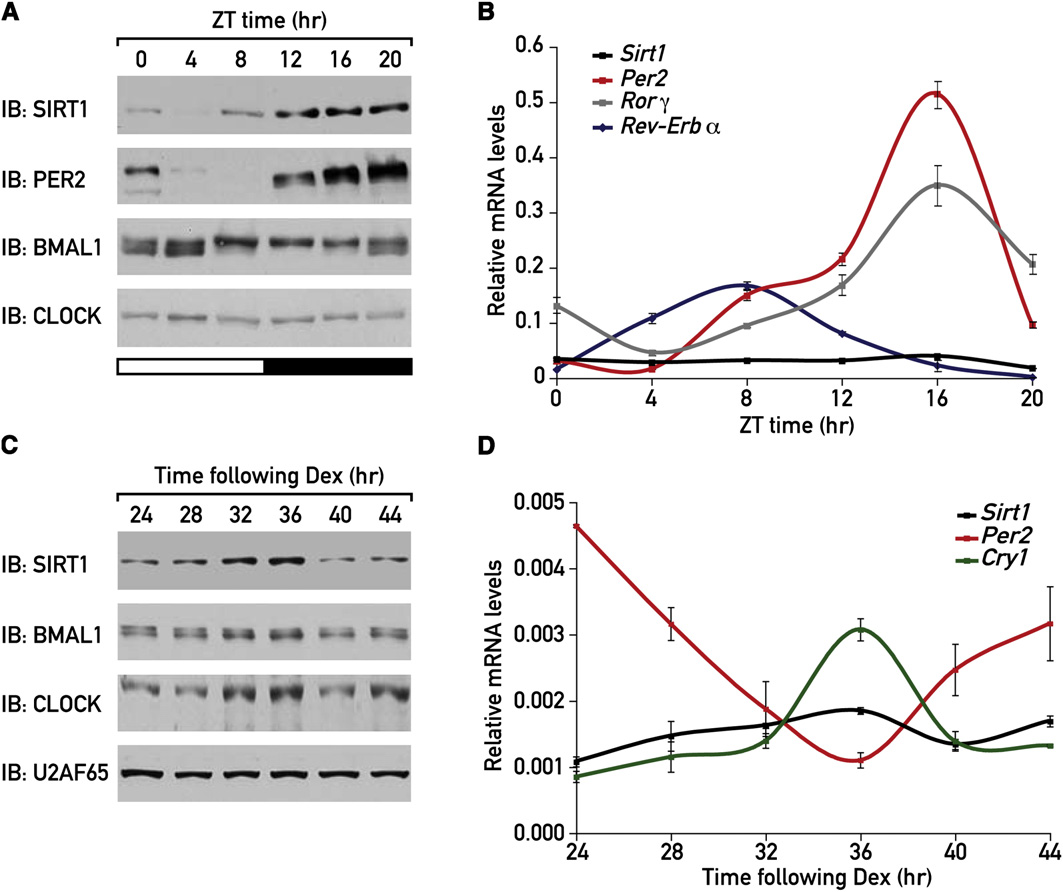
Figure 1. Circadian Expression of SIRT1 ProteinMice were sacrificed at 4 hr intervals; liver nuclear protein extracts and total RNA were prepared.
(A) Protein extracts were analyzed by immunoblotting.
(B) RNA was analyzed by quantitative TaqMan real-time PCR using specific TaqMan probes.
MEFs were synchronized by a dexamethasone shock and samples were collected at 4 hr intervals, starting 24 hr after the shock.
(C) Protein extracts were analyzed by immunoblotting.
(D) RNA was analyzed by quantitative TaqMan real-time PCR using specific TaqMan probes.
Plotted values are the mean values ± standard deviation (SD) from three independent experiments.
Here we show that SIRT1 is expressed in a circadian manner
expression as well as changes in BMAL1 phosphorylation were
and that it is required for high-magnitude circadian expression
of several core clock genes. Moreover, we present evidence
that SIRT1 binds to CLOCK-BMAL1 heterodimers and promotes
whether the daily changes in SIRT1 protein expression were
the deacetylation and degradation of PER2.
due to corresponding changes in Sirt1 mRNA accumulation,we analyzed mouse liver RNA harvested around the clock
by quantitative TaqMan real-time PCR. In contrast to severalwell-established circadian transcripts such as Per2, Rorg, and
Circadian Expression of SIRT1 Protein in Mouse Liver
Rev-Erba mRNAs, the levels of Sirt1 mRNA were nearly constant
and in Cultured Fibroblasts
throughout the day (B). Hence, posttranscriptional regu-
In order to investigate whether SIRT1 might be involved in circa-
latory mechanisms must have accounted for the observed circa-
dian rhythm, we first examined the temporal expression of SIRT1
dian changes in SIRT1 protein levels.
in mouse liver. Mice were sacrificed at 4 hr intervals around the
SIRT1 protein was also expressed in a circadian manner in
clock, and liver nuclear proteins were prepared and analyzed.
dexamethasone-synchronized cultured mouse embryonic fibro-
The results showed that SIRT1 accumulated in a circadian man-
blasts (MEFs) (and NIH 3T3 cells A available
ner with maximal and minimal levels reached at around Zeitgeber
online). Again, no significant changes in Sirt1 mRNA accumula-
time (ZT) 16 and ZT4, respectively (In parallel, we also
tion were observed As expected Per2 and
followed the temporal expression of known core clock proteins.
Cry1 mRNA levels were clearly rhythmic in these cells
As reported previously, circadian changes in PER2 protein
(Finally, we examined temporal SIRT1 accumulation
318 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
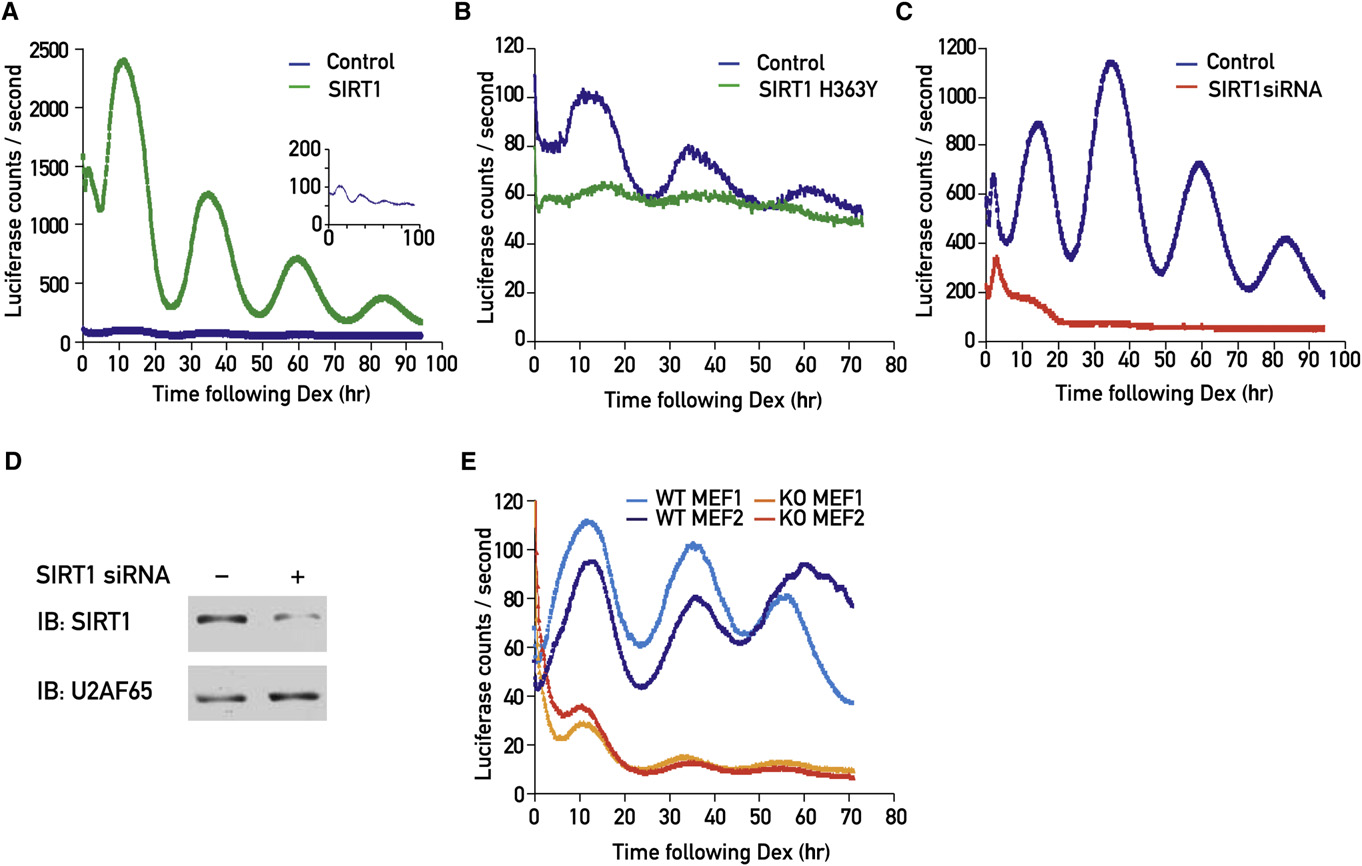
Figure 2. SIRT1 Deacetylase Activity Is Required for the High-Magnitude Oscillation of the Bmal1-Luciferase Reporter(A) NIH 3T3 cells were transfected with a 200 ng/plate Bmal1-luciferase reporter gene either alone (blue) or together with an HA-Flag-SIRT1 expression vector(green). The small insert shows the cells transfected with the Bmal1-luciferase reporter gene alone at a higher magnification.
(B) NIH 3T3 cells were transfected with a 200 ng/plate Bmal1-luciferase reporter gene either alone (blue) or together with an HA-Flag-SIRT1 H363Y expressionvector (green).
(C) NIH 3T3 cells were transfected with a 1 mg/plate Bmal1-luciferase reporter gene either with pU6 empty vector (blue) or together with pU6-Sirt1 siRNA expres-sion vector (red).
(D) Immunoblot analysis of protein extracts obtained from NIH 3T3 cells transfected with pU6 empty vector or pU6-Sirt1 siRNA expression vector.
(E) Two WT MEF cell lines (dark and light blue) and two Sirt1 KO MEF cell lines (dark and light orange), all obtained from different embryos harvested from the samepregnant female, were transduced with Bmal1-luciferase adenovirus.
Cells were synchronized by a dexamethasone shock and bioluminescence was recorded using photomultiplier tubes.
in synchronized NIH 3T3 fibroblasts by immunohistochemistry
luciferase reporter plasmid resulted in a dramatic increase in
experiments. SIRT1 staining was mostly nuclear and more in-
the magnitude of the bioluminescence oscillations (
tense at 36 hr than at 24 hr following the dexamethasone shock
The deacetylase activity of SIRT1 was required for this effect
(C and S1D). Interestingly, even in knockout (KO)
since cotransfection of SIRT1 H363Y, a catalytically inactive,
MEFs stably expressing human SIRT1 from an expression vector
dominant-negative SIRT1 mutant version
containing the SV40 promoter and unrelated 50- and 30-untrans-
virtually abolished the circadian bioluminescence oscillations
lated regions, human SIRT1 protein was expressed differentially
generated by the Bmal1-luciferase reporter gene (
at two examined time points (This experiment fa-
We also tested the effect of SIRT1 expression on reporter
vors a mechanism involving changes in protein stability rather
genes driven by other promoters, in particular Per2-luciferase,
than translation rates in circadian SIRT1 protein accumulation.
Whereas SIRT1 did not significantly affect CMV-luciferase or
SIRT1 Deacetylase Activity Is Required
Rev-Erba-luciferase expression, it increased the magnitude of
for High-Magnitude Bmal1 Expression
the bioluminescence oscillations of Per2-luciferase and Dbp-
In order to examine whether SIRT1 might influence circadian
gene expression, we followed real-time bioluminescence
Nicotinamide (NAM), a product of the SIRT1 deacetylation
recordings of a Bmal1-luciferase reporter whose expression is
reaction, was reported to inhibit SIRT1 activity (
driven by the Bmal1 promoter Cotransfec-
Treatment of NIH 3T3 cells stably expressing the
tions of a SIRT1 expression vector together with the Bmal1-
Bmal1-luciferase reporter gene with a moderate concentration
Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 319
of NAM (10 mM) had no significant effect on the daily biolumines-
proteins for ubiquitination and degradation), and Cki3 (casein
cence oscillations A higher concentration of NAM
kinase I3) were only slightly affected by the absence of SIRT1
(50 mM) resulted in the dampening of circadian Bmal1-luciferase
(The strong repression of Rorg mRNA expression in
expression A). However, the most conspicuous effect
the Sirt1 KO MEFs incited us to examine the levels of the other
of NAM treatment was a dramatic period lengthening
two Ror paralogs, Rora and Rorb . However, no significant
a phenotype that was not observed in genetic loss-
differences were observed for Rora mRNA levels, and Rorb, a
of-function experiments (see below) and, therefore, was unlikely
neuron-specific Ror isoform ), was unde-
to involve SIRT1. In contrast, Sirtinol, a more specific and potent
tectable in both WT and Sirt1 KO MEFs (
inhibitor of SIRT1 deacetylation activity (),
We also examined the accumulation of various clock proteins
strongly dampened the circadian Bmal1-luciferase reporter
from nonsynchronized WT and Sirt1 KO MEFs. In keeping with
gene expression (B) and thereby closely phenocopied
the changes observed for their mRNA levels, both BMAL1 and
the phenotypes observed in genetic Sirt1 loss-of-function
CLOCK protein levels were significantly downregulated in the
Sirt1 KO MEFs However, although the Per2 mRNA
To further scrutinize possible roles of SIRT1 on the clock func-
level was strongly decreased in Sirt1 KO MEFs (
tion we performed loss-of-function experiments. Cotransfection
PER2 protein accumulation was actually higher in these cells
of a Sirt1 siRNA expression vector with the Bmal1-luciferase
(C). CRY1 protein accumulation was also slightly ele-
reporter gene attenuated the circadian oscillations (
vated in the absence of SIRT1 C), in contrast to the
in a dose-dependent manner (The effect of Sirt1
downregulation of its mRNA level (
siRNA expression on endogenous SIRT1 protein accumulation
To verify whether the described changes in clock gene expres-
was verified by analysis of protein extracts from the cells that
sion were indeed due to the absence of SIRT1 in the KO MEFs,
were used for the Bmal1-luciferase recordings (D). Since
we generated rescue cell lines from Sirt1 KO MEFs that stably
not all cells contributing to the protein extract were transfected,
expressed a human SIRT1 cDNA. The comparison of WT, Sirt1
the downregulation of SIRT1 accumulation in transfected cells
KO, and two human SIRT1-rescued KO MEFs cell lines showed
must have been very efficient. The expression of Sirt1 siRNA
that the mRNA levels encoding the different clock transcription
also resulted in a decrease in the bioluminescence oscillations
factors and human SIRT1 were restored to nearly normal levels
of Per2-luciferase and to a lesser extent of Dbp-luciferase oscil-
in both rescued cell lines (D). Similarly, the magnitude
lations and S4C) but did not affect the biolumines-
of the bioluminescence oscillations of Bmal1-luciferase re-
cence of CMV-luciferase or Rev-Erba-luciferase (D
porter were reestablished in the human SIRT-rescued KO
and S4E). We also examined the effect of SIRT1 knockdown
on Bmal1-luciferase reporter at a single-cell resolution. As can
The observed changes in clock gene mRNA and protein levels
be concluded from a comparison of (control) and
were obtained in nonsynchronized cells and thus reflected
(Sirt1 siRNA) only a small proportion of cells displayed strong
average expression levels. Thus, we also wished to monitor
bioluminescence cycles when Sirt1 expression was diminished.
the circadian expression of these genes in synchronized MEFs.
Experiments with MEFs from wild-type (WT) and Sirt1 KO em-
In accordance with the observations made in the experiments
bryos, transduced with an adenoviral vector harboring the
with the Bmal1-luciferase reporter (E), endogenous
Bmal1-luciferase reporter gene, substantiated the observations
Bmal1 mRNA was expressed at low and nearly invariable levels
made with Sirt1 siRNA-expressing cells. Thus, the magnitude
throughout the day A). Similarly, Rorg mRNA accumula-
of bioluminescence cycles was considerably higher in WT
tion was strongly repressed in the absence of SIRT1
MEFs than in Sirt1 KO MEFs (E). A closer inspection of
Per2 and Cry1 mRNA were still expressed in a circadian manner
our loss-of-function data also revealed a modest phase advance
in Sirt1 KO MEFs, but with a significantly reduced magnitude
for the temporal expression of Bmal1-luciferase, Per2-luciferase,
(Again, no significant changes in Rev-Erba and Dbp
and Dbp-luciferase in the absence of SIRT1 (E and ).
mRNA accumulation were noticed These results
However, in none of these experiments were significant differ-
were thus in keeping with the changes observed in our analysis
ences in period lengths noticed.
of nonsynchronized MEFs (
Similarly to the changes observed for Bmal1 mRNA expres-
SIRT1 Influences the Expression of Endogenous
sion A), BMAL1 protein levels were significantly downre-
gulated in the Sirt1 KO MEFs B). In contrast, both PER2
To examine whether SIRT1 also affected the expression of en-
and CRY1 protein levels were elevated and relatively constant in
dogenous circadian genes, we analyzed the levels of various
the absence of SIRT1 (B), in spite of their diminished
transcripts in WT and Sirt1 KO MEFs by quantitative TaqMan
mRNA levels Again, SIRT1 accumulated in a circa-
real-time PCR. The levels of endogenous Bmal1 mRNA were
dian manner with maximal expression between 32 to 36 hr
reduced to around 40% in nonsynchronized Sirt1 KO MEFs
following the dexamethasone shock B).
compared to WT MEFs Clock, Per1, and Cry1mRNA accumulation was attenuated to a similar extent in Sirt1
SIRT1 Binds to CLOCK-BMAL1 and PER2
KO MEFs (A), while Per2 and Rorg transcript levels in
in a Circadian Fashion
Sirt1 KO cells only amounted to 20% and 10%, respectively, of
SIRT1 was previously reported to deacetylate several tran-
those observed in WT cells (In contrast, the mRNA
scriptional regulatory proteins
levels of Rev-Erba, Dbp, bTrcp (an F box protein targeting PER
including the basic helix-loop-helix
320 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
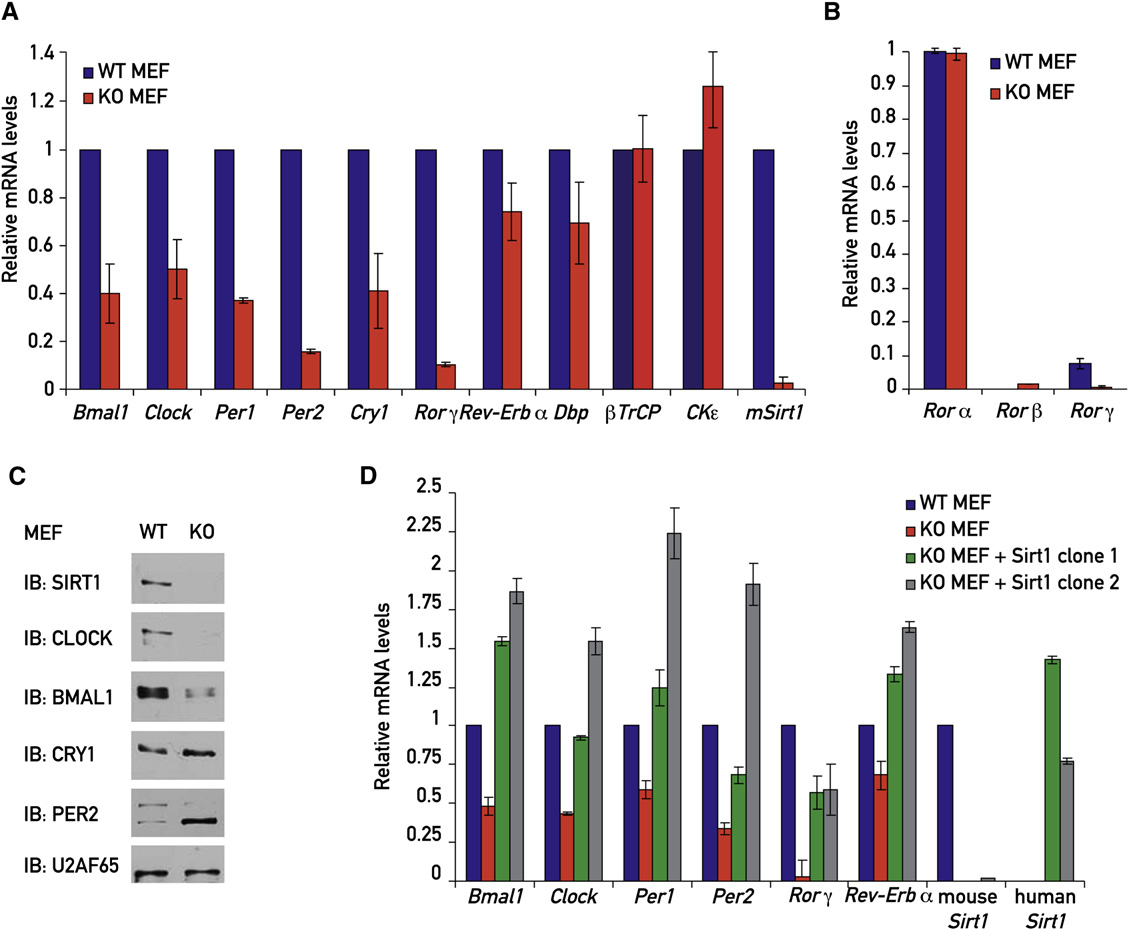
Figure 3. Analysis of mRNA and Protein Levels of Core Clock Proteins in Nonsynchronized WT, Sirt1 KO, and Sirt1 Rescue MEFs(A) RNA extracts from nonsynchronized WT and Sirt1 KO MEFs were analyzed by quantitative TaqMan real-time PCR using specific TaqMan probes.
(B) RNA extracts from nonsynchronized WT and Sirt1 KO MEFs were analyzed for the mRNA expression of different ROR isoforms by quantitative TaqMan real-time PCR.
(C) Protein extracts from nonsynchronized WT and Sirt1 KO MEFs were examined by immunoblotting.
(D) RNA extracts from nonsynchronized WT, Sirt1 KO, and two different monoclonal Sirt1 rescue MEF lines expressing human SIRT1 were analyzed by quan-titative TaqMan real-time PCR using specific TaqMan probes.
Plotted values are the mean values ± SD from three independent experiments.
repressors HES1 and HEY2 Since,
observed around ZT0 After a longer exposure
similarly to HES1 and HEY2, both CLOCK and BMAL1 contain
weak binding of PER2 to CLOCK was also detected around
helix-loop-helix domains, we first examined whether SIRT1
ZT4 (data not shown). Importantly, an immunoprecipitation ex-
might interact with these transcription factors. Both endogenous
periment with SIRT1 antibody with the same protein extracts
CLOCK and BMAL1 coimmunoprecipitated with SIRT1 in ex-
showed that SIRT1 bound to CLOCK in a circadian manner
tracts obtained from mouse liver nuclei (and from cul-
with maximal binding around ZT4 F). A similar experi-
tured NIH 3T3 fibroblasts In addition, coimmunos-
ment was conducted with whole-cell extracts from NIH 3T3
taining experiments for SIRT1 and CLOCK in NIH 3T3 cells
cells synchronized by a dexamethasone shock. The binding of
showed that at least a fraction of these proteins colocalized in
CLOCK-BMAL1 to SIRT1 was circadian with maximal binding
between 36 and 42 hr after the dexamethasone treatment
The binding of SIRT1 to CLOCK and BMAL1 prompted us to
(Interestingly, PER2 could also be detected in
examine whether SIRT1 interacted with additional core clock
SIRT1-associated complexes around 42 hr after the dexameth-
proteins in a circadian manner. We thus analyzed the different
asone shock.
binding partners of CLOCK around the clock in mouse liver nu-clear extracts (As expected, immunoprecipitaiton of
SIRT1 Deacetylates PER2
CLOCK resulted in the coimmunoprecipitation of BMAL1, with
To examine whether CLOCK, BMAL1, or PER2 are acetylated
maximal binding around ZT8 E). In contrast, maximal in-
and thereby potential substrates for SIRT1 deacetylation
teractions of the repressors PER2 and CRY1 with CLOCK were
activity, we performed immunoprecpitation experiments with
Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 321
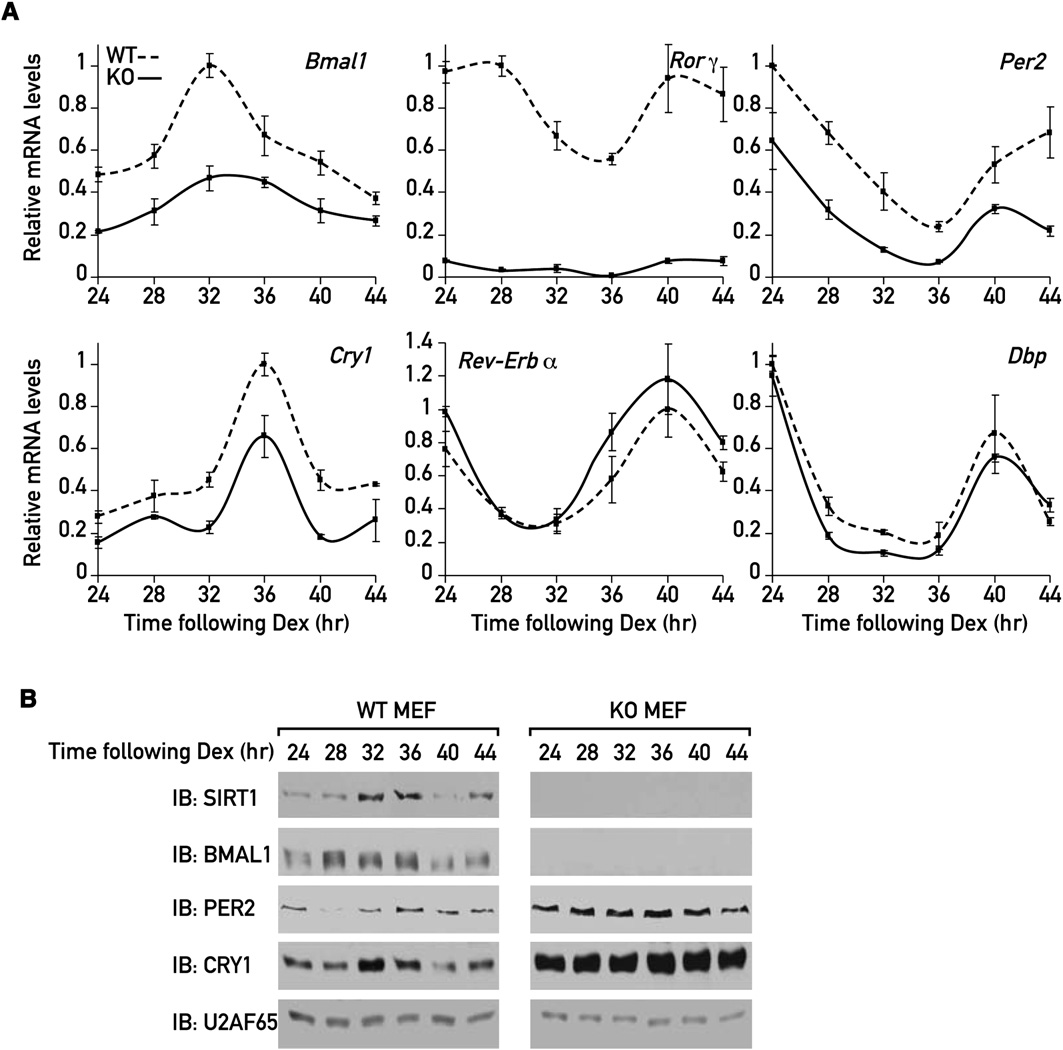
Figure 4. Analysis of Circadian mRNA and Protein Levels of Core Clock Proteins in WT and Sirt1 KO MEFsWT and Sirt1 KO MEFs were synchronized by a dexamethasone shock, and protein and RNA were extracted at 4 hr intervals starting 24 hr after the shock.
(A) mRNA analysis was done by quantitative TaqMan real-time PCR using specific TaqMan probes. Plotted values are the mean values ± SD from three inde-pendent experiments.
(B) Protein extracts were analyzed by immunoblotting.
nonsynchronized WT and Sirt1 KO MEFs, using a pan acetyl ly-
itation experiments were performed with a pan acetyl lysine an-
sine antibody. The immunoprecipitated proteins were analyzed
tibody, and the immunoprecipitated proteins were analyzed by
by immunoblotting using antibodies for various clock proteins.
immunoblotting. In agreement with the previous experiments,
These experiments failed to reveal acetylated forms of CLOCK,
no acetylation of BMAL1 or CLOCK was detected
BMAL1, CRY1, or SIRT1 but suggested that a frac-
In WT MEFs, PER2 acetylation was maximal at around 32 hr after
tion of PER2 was acetylated in Sirt1 KO MEFs Low
the dexamethasone shock (C). The extent of PER2 acet-
levels of acetylated PER2 were also detected in WT MEFs after
ylation was significantly higher in KO MEFs than in WT MEFs,
a long exposure (data not shown). To corroborate PER2 acety-
and maximal acetylation in KO MEFs occurred between 24 and
lation we transfected NIH 3T3 cells with expression vectors for
28 hr after the dexamethasone shock. Our findings thus sug-
a tandem affinity purification (TAP) tagged PER2 (PER2-TAP)
gested that PER2 was acetylated in a circadian manner, and
or a TAP-tagged luciferase (luciferase-TAP) (as a negative
that SIRT1 deacetylated PER2 in vivo. While PER2 protein levels
control). Immunoblot analysis of purified TAP-tagged proteins
were elevated and relatively constant in the Sirt1 KO MEFs
confirmed that PER2 was acetylated ) and that the
(PER2 acetylation and/or deacetylation was cyclic.
acetyl groups were removed in vitro by recombinant SIRT1 in
This is in line with the circadian binding of SIRT1 to PER2
an NAD+-dependent manner B).
(To examine whether PER2 was also acetylated in
Next, we monitored the acetylation of PER2 in synchronized
mice, we immunoprecipitated PER2 from mouse liver nuclear
WT and Sirt1 KO MEFs around the clock. Again, immunoprecip-
extracts prepared around the clock. Immunoblot analysis of
322 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
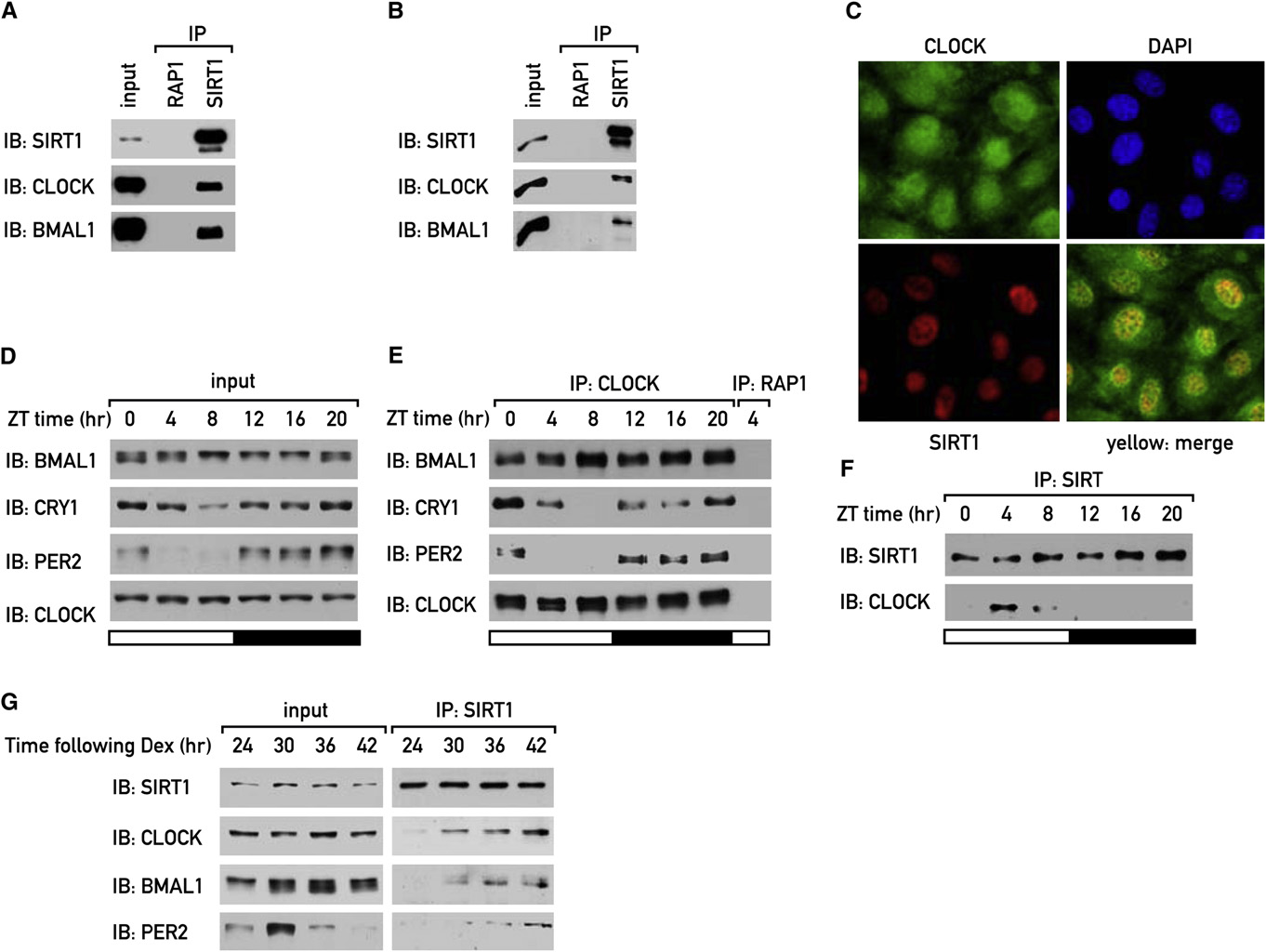
Figure 5. SIRT1 Binds to CLOCK, BMAL1, and PER2 in a Circadian MannerSIRT1 was immunoprecipitated from mouse liver nuclear extracts (A) and from NIH 3T3 cells (B). The immunoprecipitated proteins were analyzed by immuno-blotting. Rabbit yeast RAP1 antibody was used as a negative control.
(C) Immunostaining of SIRT1 (red) and CLOCK (green) in NIH 3T3 cells was performed with rabbit SIRT1 and CLOCK antibodies. In blue: DAPI staining. In yellow:merge of SIRT1 and CLOCK staining.
(D) Mice were sacrificed at 4 hr intervals, and liver nuclear extracts were analyzed by immunoblotting.
(E) CLOCK was immunoprecipitated from mouse liver nuclear extracts, and the immunoprecipitated proteins were analyzed by immunoblotting. Rabbit yeastRAP1 antibody was used as a negative control.
(F) SIRT1 was immunoprecipitated from mouse liver nuclear extracts, and the immunoprecipitated proteins were analyzed by immunoblotting.
(G) NIH 3T3 cells were synchronized by a dexamethasone shock, and protein extracts were prepared at 6 hr intervals, starting 24 hr after the shock. SIRT1 wasimmunoprecipitated from NIH 3T3 cells, and the immunoprecipitated proteins were analyzed by immunoblotting.
precipitated proteins with the pan acetyl lysine antibody showed
during 4 hr. The results indicated that the protein half-life of
that PER2 was acetylated also in mouse liver D).
PER2 was significantly prolonged in the absence of SIRT1(and 7B). In keeping with these observations, the
SIRT1-Dependent Deacetylation of PER2 Determines
coexpression of SIRT1 together with PER2-TAP significantly
PER2 Protein Stability
reduced the accumulation C) and acetylation of PER2-
The elevated PER2 acetylation and accumulation in Sirt1 KO
TAP (whereas knockdown of SIRT1 expression with
MEFs on one hand and the reduced Per2 mRNA levels on the
Sirt1 siRNA resulted in a significant increase in PER2 accumula-
other hand raised the possibility that acetylation of PER2 stabi-
tion and acetylation E).
lized the protein. We thus compared the decay of PER2 protein
To further address the dependency of PER2 degradation upon
in the presence and absence of SIRT1. To this end, WT and Sirt1
deacetylation by SIRT1 we performed an in vitro assay with
KO MEFs were incubated with or without cycloheximide 24 hr
purified PER2-TAP and extracts obtained from WT and Sirt1 KO
after synchronization and PER2 protein levels were recorded
MEFs. PER2 was deacetylated only in the presence of extracts
Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 323
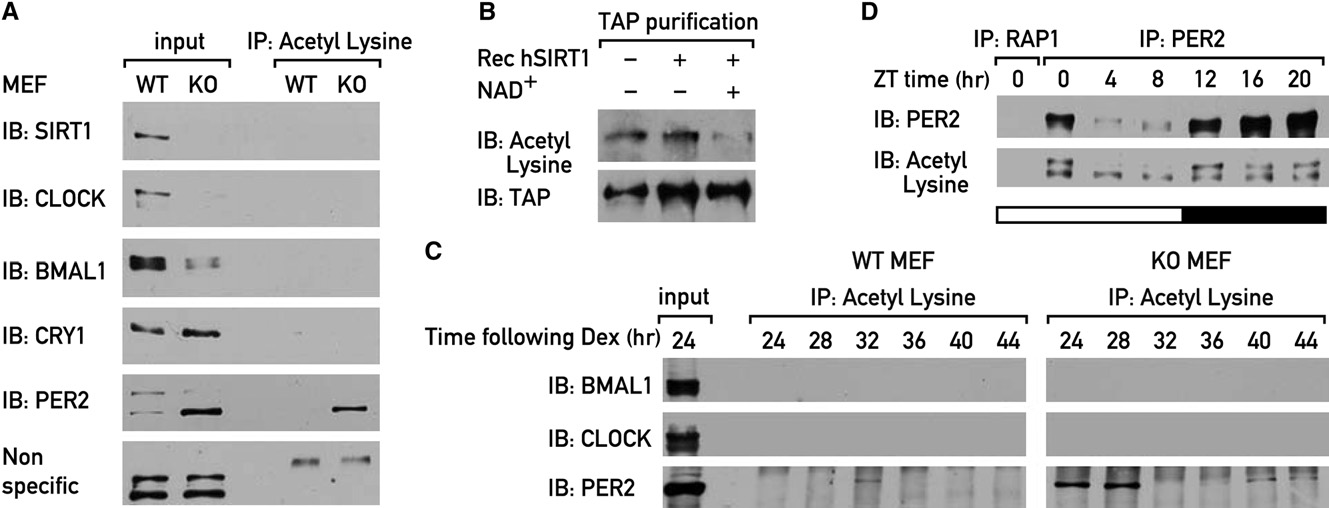
Figure 6. SIRT1 Deacetylates PER2(A) Protein extracts from nonsynchronized WT and Sirt1 KO MEFs were subjected to immunoprecipitation with rabbit pan acetyl lysine antibody, and the immu-noprecipitated proteins were analyzed by immunoblotting.
(B) Purified PER2-TAP was incubated in the absence or presence of recombinant SIRT1 and NAD+ for 3 hr at 30�, and samples were analyzed by immunoblotting.
(C) WT and Sirt1 KO MEFs were synchronized by a dexamethasone shock, and protein extracts were prepared at 4 hr intervals, starting 24 hr after the shock.
Immunoprecipitation experiments were performed with rabbit pan acetyl lysine antibody, and the immunoprecipitated proteins were analyzed by immunoblotting.
(D) Mice were sacrificed at 4 hr intervals and liver nuclear extracts were prepared. PER2 was immunoprecipitated, and the immunoprecipitated proteins wereanalyzed by immunoblotting. Rabbit yeast RAP1 antibody was used as a negative control.
from WT MEFs together with NAD+. Deacetylation of PER2 re-
tained from wild-type but not from Sirt1 KO MEFs, and recombi-
sulted in PER2 degradation, which was blocked in the presence
nant SIRT1 deacetylates purified PER2 in vitro in a NAD+-depen-
of the proteasome inhibitor MG132
dent manner. The latter result should, however, be interpretedwith caution since the in vitro substrate specificity of recombi-
nant SIRT1 is rather promiscuous ().
Other posttranslational modifications such as phosphoryla-
Modulation of Circadian Oscillator Function by Protein
tion, sumoylation, histone acetylation, and methylation have
Acetylation and Deacetylation
already been shown to play a key role in circadian gene expres-
We identified SIRT1 as a regulator of circadian gene expression.
sion ). For example, sumoylation of
SIRT1 accumulates in a circadian manner in mouse hepatocytes
BMAL1 has been shown to play an important role in BMAL1 ac-
and cultured fibroblasts and is required for high-magnitude cir-
cumulation and clock rhythmicity (Like-
cadian transcription of several core clock genes, including
wise, phosphorylation of BMAL1 either by Casein Kinase I (CKI)
Bmal1, Rorg, Per2, and Cry1. SIRT1 binds to CLOCK-BMAL1
(or by mitogen-activated protein kinases
and PER2 in a circadian manner and supports the deacetylation
(MAPK) () modulates BMAL1-CLOCK-depen-
and degradation of PER2. In the absence of SIRT1, constitutively
dent transcription. Recently, CLOCK was reported to acetylate
high protein levels of PER2 may lead to the repression of Per1,
BMAL1, thereby facilitating repression of BMAL1-CLOCK-de-
Per2, Cry1, and Rorg mRNA expression. Repression of RORg,
pendent transcription We suppose that
an activator of Bmal1 transcription, is likely to account for the
due to the sensitivity of our immunoblot experiments, the acety-
dampening of Bmal1 mRNA and protein expression in Sirt1 KO
lated fraction of endogenous BMAL1 in liver and fibroblast ex-
tracts was not revealed. CKI has been reported to phosphorylate
The enzyme(s) responsible for PER2 acetylation remain(s) to
PER2 protein, thereby regulating PER2 protein stability (
be identified, but the acetyltransferase activity of CLOCK
). Likewise, CRY degradation mediated by the F box
or p300, a coactivator associated with CLOCK-
protein SCFFbxl3 has been demonstrated to be required for
BMAL1 heterodimer (are attractive can-
normal oscillator function
didates. PGC1a, a recently identified key player in circadian
oscillator function ), may also affect PER2 acety-lation via stimulating the acetyl transferase activity of p300
SIRT1 Affects Circadian Transcription
in a Gene-Specific Manner
Although our results cannot rigorously exclude a more compli-
The extent to which SIRT1 affects circadian transcription ap-
cated scenario, they suggest that SIRT1 deacetylates PER2
pears to be target gene specific. For example, in the absence
directly. Thus, SIRT1 is associated with CLOCK-BMAL1-PER2
of SIRT1, Rorg mRNA levels are strongly repressed and Per2
complexes, purified PER2 is deacetylated in vitro by extracts ob-
mRNA levels are significantly downregulated, whereas Rev-Erba
324 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
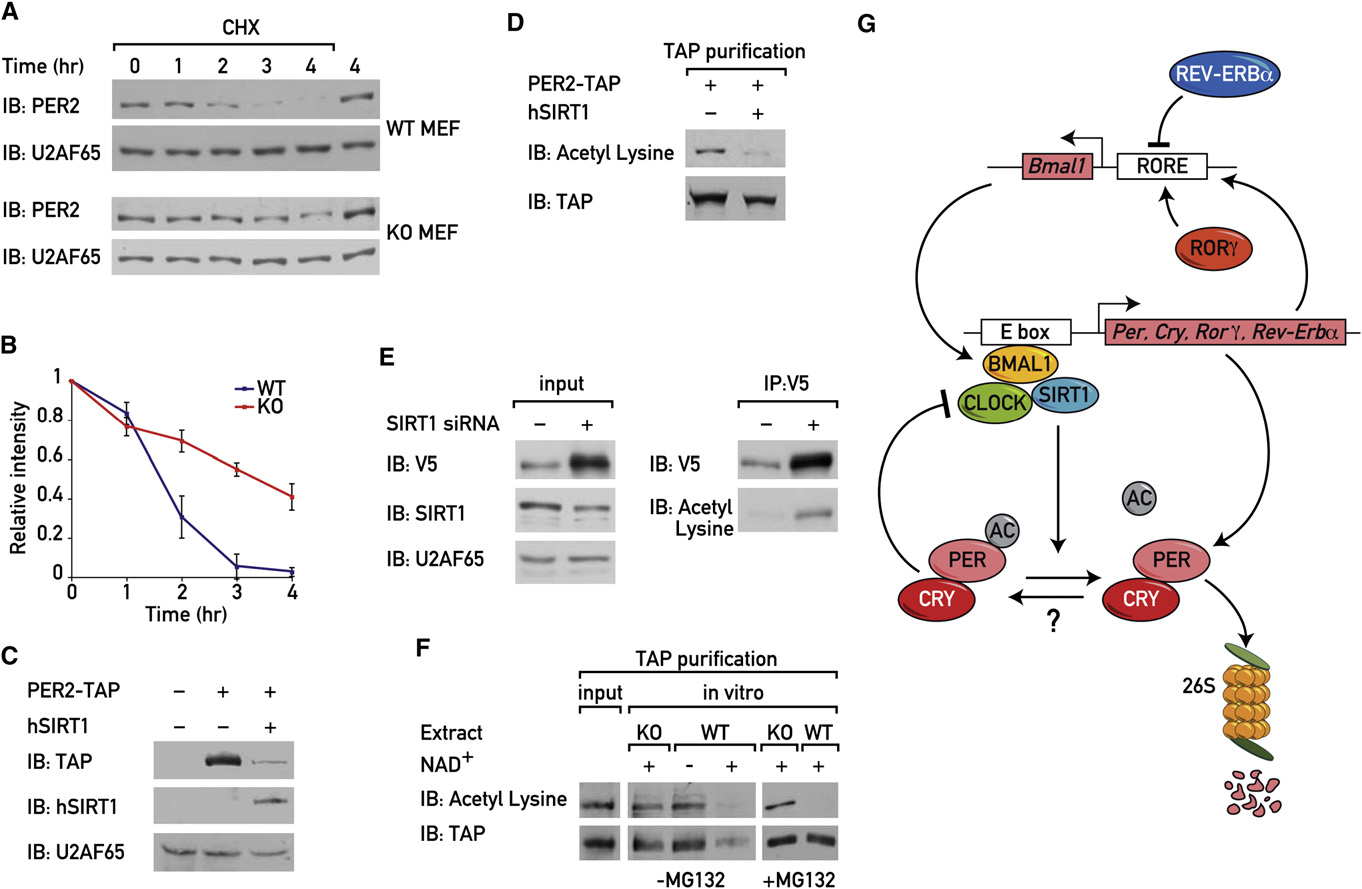
Figure 7. SIRT1-Dependent PER2 Deacetylation Determines PER2 Protein Stability(A) WT and Sirt1 KO MEFs were synchronized by a dexamethasone shock, and 24 hr after the shock cells were untreated or treated with cycloheximide. Cellswere harvested 1, 2, 3, and 4 hr following the treatment, and protein extracts were analyzed by immunoblotting.
(B) The graph illustrates the quantification of PER2 by densitometry of triplicate experiments (mean ± standard error).
(C) NIH 3T3 cells were transfected with PER2-TAP expression vector either alone or together with HA-FLAG-human SIRT1 expression vector. Protein extractswere analyzed by immunoblotting.
(D) PER2-TAP was purified from NIH 3T3 cells transfected with PER2-TAP expression vector either alone or together with HA-FLAG-human SIRT1 expressionvector and analyzed by immunoblotting.
(E) NIH 3T3 cells were transfected with the V5-PER2 expression vector either alone or together with the Sirt1 siRNA expression vector. Protein extracts wereprepared, and immunoprecipitation experiments were performed with mouse V5 antibody. The immunoprecipitated proteins were analyzed by immunoblotting.
(F) Purified PER2-TAP was incubated for 3 hr at 30� with protein extract obtained from WT or Sirt1 KO MEFs in the absence or presence of 100 mM NAD+ or 25 mMMG132, and samples were analyzed by immunoblotting.
(G) Hypothetical model showing the possible role of SIRT1 in circadian oscillator function. BMAL1-CLOCK heterodimers bind and activate transcription of the Per,Cry, Rre-Erba, and Rorg genes. Once the PER and CRY proteins accumulate to a critical level, they form complexes with BMAL1-CLOCK and thereby represstheir own transcription. In addition, there is an interconnecting feedback loop in which REV-ERBa represses and RORg activates Bmal1 transcription. SIRT1binds CLOCK-BMAL1 complexes and promotes PER2 deacetylation and degradation.
and Dbp mRNA levels are only slightly affected Con-
SIRT1 is caused by a combination of diminished RORg
ceivably, BMAL1 and CLOCK bind their DNA cognate sites in
expression and impaired PGC1a coactivation. Indeed, Pgc1a
Rev-Erba and Dbp with a higher affinity than those present in
KO mice exhibit abnormal diurnal rhythms of activity, body
Per2 and Rorg. The reduced BMAL1-CLOCK levels in Sirt1 KO
temperature, and metabolic rate (Unfortu-
cells might then still support high-amplitude/magnitude Rev-
nately, such studies cannot be performed with Sirt1-deficient
Erba and Dbp transcription.
mice because their postnatal survival rates are very poor and
PGC1a was found to be expressed in a circadian manner
the few surviving mice exhibit many developmental defects
and to stimulate Bmal1 transcription as a coactivator of the
ROR family of nuclear orphan receptors ).
SIRT1 deacetylates PGC1a and thereby modulates its coacti-
Regulation of PER2 Protein Degradation
vator activity (). Thus, it is possible that
In mammals, the stability of PER proteins is regulated by
the downregulation of Bmal1 expression in the absence of
the F-box-containing E3 ubiquitin ligase bTrCP (
Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 325
). PER phosphorylation by CKI3 promotes the re-
Plasmids and Transfections
cruitment of bTrCP complexes, which in turn mediates the ubiq-
The following plasmids were used: Bmal1-luciferase ),
uitination and proteasomal degradation of PER
Per2(E-BOX2)-luciferase (), Dbp-luciferase (), Rev-Erba-luciferase, and CMV-luciferase; pCDNA HA-Flag-SIRT1 en-
Our results that both PER2 and acety-
coding WT human SIRT1, pCDNA HA-Flag-SIRT1 H363Y encoding inactive
lated PER2 levels are elevated in the absence of SIRT1 suggest
deacetylase mutant of SIRT1, pBabe human SIRT1, pU6-siRNA-Sirt1, and
that SIRT1-mediated deacetylation enhances the rate of PER2
pU6-empty vector (and pEF5/FRT7V5-PER2, pCMV
degradation. Since ubiquitination and acetylation occur on lysine
PER2-TAP, and pCMV luciferase-TAP. Transient transfections of NIH 3T3 cells
residues, it is conceivable that the same lysine residues can be
were carried out with FuGENE Transfection Reagent (Roche) according to the
either acetylated or ubiquitinated. If true, acetylated PER2 could
no longer be ubiquitinated and degraded by the proteasome,
Generation and Transduction of Bmal1-Luciferase
which would explain the augmented PER2 levels in Sirt1 KO
Expressing Adenovirus
cells. Interestingly, CRY1 protein levels are also elevated in the
Bmal1-luciferase (cassettes was cloned into pCV100
absence of SIRT1, in spite of reduced Cry1 mRNA levels
plasmid, first-generation adenoviral vector was amplified in N52.E6-producer
). It has been reported that PER2 inhibits the ubiquitination
cells, and viruses were generated and purified as previously described (
and degradation of CRY proteins ). Thus, the
MEFs were incubated with Bmal1-luciferase expressing adenovirus at an
higher accumulation of PER2 in Sirt1 KO cells may account for
multiplicity of infection (moi) of 5000 for 6 hr, cells were thoroughly washed
the elevated CRY1 levels in these cells.
with PBS, and the medium was replaced. Forty-eight hours after transductioncells were shocked with dexamethasone and real-time bioluminescence was
The Circadian Accumulation and Activity
Circadian SIRT1 protein accumulation appears to be con-
Cells were fixed with 4% paraformaldehyde for 10 min at room temperature.
trolled by posttranscriptional mechanisms as no significant
Fixed cells were permeabilized with 0.5% Triton X-100 in TBS and washed
changes in Sirt1 mRNA were observed ). Surprisingly,
with TBS containing 0.1% Triton X-100 (TBS-T). Samples were blocked with
temporal SIRT1 accumulation does not correlate with the cir-
2% BSA in TBS-T followed by incubation with rabbit anti-SIRT1 and rat anti-
cadian interaction of SIRT1 with its identified core clock bind-
CLOCK antibodies. Samples were washed with TBS-T and incubated withalexa 594-conjugated anti-rabbit and with FITC-conjugated anti-rat secondary
ing partners. For example, in mouse liver nuclei, maximal
antibodies. Nuclei were stained with DAPI. Microscopic images were obtained
SIRT1 protein expression is observed at around ZT16,
using a Leica SP2 confocal microscope.
whereas its maximal binding to CLOCK occurs at aroundZT4, when SIRT1 levels are minimal. Similarly, in NIH 3T3
RNA Analysis by Real-Time Quantitative PCR
cells, maximal SIRT1 protein expression occurs between 32
RNA extraction and transcript quantification by TaqMan real-time PCR tech-
and 36 hr after the dexamethasone shock, while its maximal
nology was performed as previously described ), usingan ABI PRISM 7700 Sequence Detection System from PE-Applied Biosys-
binding to the core clock components is observed around
tems. The real-time PCR data were normalized to 45S pre-mRNA. Primers
42 hr. Therefore, SIRT1 might also regulate the expression
and probes are listed in
of circadian output genes expressed with a different phase,possibly through activation of coactivators and transcription
Protein Extraction and Immunoblot Analysis
factors such as PGC1a, FOXO, or LXR
Proteins from mouse liver nuclei and cultured fibroblasts were prepared
) or through the circadian deacetylation
according to the NUN procedure (Trichostatin A
of histones in nucleosomes associated with clock-controlled
was added during the extraction. SDS-PAGE and immunoblot analysis wereperformed according to standard protocols. Antibodies used were rabbit
CRY1, PER2, BMAL1, and CLOCK (kindly provided by S. Brown and J. Rip-
perger) and rabbit SIRT1 (Upstate), human-SIRT1 (Santa Cruz), pan acetylated
Future experiments with Sirt1-deficient and -proficient
lysine (Cell Signaling), TAP (OPEN BIOSYSTEMS), mouse V5 (Invitrogen), and
cells should shed light on the role of SIRT1 in these additional
U2AF65 (Sigma).
mechanisms involved in the regulation of circadian geneexpression.
Coimmunoprecipitation experiments were carried out with mouse liver nuclearextracts or with whole-cell NUN extracts. Extracts were incubated for 12 hr
EXPERIMENTAL PROCEDURES
with the indicated antibodies at 4�C and further incubated with protein A beads(Roche) for an additional 2 hr at 4�C. The beads were collected by centrifuga-
Cells and Cell Culture
tion and washed with NP40 buffer (100 mM Tris-HCL pH 7.5, 150 mM NaCl,
NIH 3T3 cells and NIH 3T3-Bmal1-luciferase cells stably expressing Bmal1-lu-
2 mM EDTA, and 1% NP40). Laemmli sample buffer was added and samples
ciferase reporter were grown as previously described (
were heated at 95�C for 5 min and loaded on a polyacrylamide-SDS PAGE.
WT and Sirt1 KO MEFs were grown in Dulbecco's modified Eagle's medium
Purification of the C-terminal TAP-tagged PER2 and TAP-tagged luciferase
(DMEM) supplemented with 15% FBS, 100 units/ml penicillin, 100 mg/ml
proteins were performed according to standard protocol as previously
streptomycin, 2 mM glutamine, 8 mM nonessential amino acids (Sigma),
described ().
1 mM Na-Pyruvate, 0.006 mM b-mercaptoethanol, and 18 mM HEPES(pH 7.0) and cultured at 37�C in a humidified incubator with 5.6% CO2. Cells
In Vitro Deacetylation Assay
were synchronized with 100 nM dexamethasone and real-time biolumines-
Purified PER2-TAP protein was incubated in deacetylation buffer (50 mM Tris-
cence was recorded ). Nicotinamide and dexamethasone
HCL pH 8, 50 mM NaCl, 4 mM MgCl2) in the presence of purified recombinant
were prepared in H2O and ethanol, respectively. Trichostatin a, cycloheximide,
human SIRT1 (BioMol, 5U) or in the presence of protein extracts from WT or
and sirtinol (Sigma) were dissolved in DMSO.
Sirt1 KO MEFs lysed in RIPA lysis buffer (150 mM NaCl, 1% NP-40 [vol/vol],
326 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
0.5% Na-deoxycholate [DOC vol/vol], 0.1% SDS [vol/vol], 50 mM Tris-Hcl
the circadian clock by directing the degradation of cryptochrome proteins.
pH 8, 1 mM dithiothreitol [DTT]). Reactions were carried out in the presence
Science 316, 900–904.
or absence of 100 mM NAD+ for 3 hr at 30�.
Cardone, L., Hirayama, J., Giordano, F., Tamaru, T., Palvimo, J.J., andSassone-Corsi, P. (2005). Circadian clock control by SUMOylation of
SUPPLEMENTAL DATA
BMAL1. Science 309, 1390–1394.
Cheng, H.L., Mostoslavsky, R., Saito, S., Manis, J.P., Gu, Y., Patel, P., Bron-
Supplemental Data include six figures, one table, and two movies and can be
son, R., Appella, E., Alt, F.W., and Chua, K.F. (2003). Developmental defects
found with this article online at
and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl.
Acad. Sci. USA 100, 10794–10799.
Cohen, H.Y., Miller, C., Bitterman, K.J., Wall, N.R., Hekking, B., Kessler, B.,
Howitz, K.T., Gorospe, M., de Cabo, R., and Sinclair, D.A. (2004). Calorie re-striction promotes mammalian cell survival by inducing the SIRT1 deacetylase.
We thank S. Brown and J. Ripperger for the CRY1, PER2, BMAL1, and CLOCK
Science 305, 390–392.
antibodies; A. Kramer, J. Takahashi, and J. Ripperger for the pEF5/FRT7V5-
Curtis, A.M., Seo, S.B., Westgate, E.J., Rudic, R.D., Smyth, E.M., Chakravarti,
PER2, Per2(E-BOX2)-luciferase, and Rev-Erba-luciferase plasmids, respec-
D., FitzGerald, G.A., and McNamara, P. (2004). Histone acetyltransferase-de-
tively; D. Welsh for advice on single-cell luminescence imaging; and N. Roggi
pendent chromatin remodeling and the vascular clock. J. Biol. Chem. 279,
for the artwork. This research was supported by the Swiss National Foundation
(through an individual research grant to U.S. and the National Center of Com-petence in Research Program Frontiers in Genetics), the State of Geneva, the
Dali-Youcef, N., Lagouge, M., Froelich, S., Koehl, C., Schoonjans, K., and
Louis Jeantet Foundation of Medicine, the Bonizzi-Theler Stiftuung, and the 6th
Auwerx, J. (2007). Sirtuins: The ‘magnificent seven', function, metabolism
European Framework Project EUCLOCK. G.A. and H.R. received long-term
and longevity. Ann. Med. 39, 335–345.
fellowships from EMBO and Human Frontier Science Program. D.G. received
Damiola, F., Le Minh, N., Preitner, N., Kornmann, B., Fleury-Olela, F., and Schi-
long-term fellowships from FEBS and Human Frontier Science Program.
bler, U. (2000). Restricted feeding uncouples circadian oscillators in peripheraltissues from the central pacemaker in the suprachiasmatic nucleus. Genes
Received: October 24, 2007
Dev. 14, 2950–2961.
Revised: March 17, 2008
DeBruyne, J.P., Weaver, D.R., and Reppert, S.M. (2007). CLOCK and NPAS2
Accepted: June 23, 2008
have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci.
Published: July 24, 2008
Doi, M., Hirayama, J., and Sassone-Corsi, P. (2006). Circadian regulator
CLOCK is a histone acetyltransferase. Cell 125, 497–508.
Akhtar, R.A., Reddy, A.B., Maywood, E.S., Clayton, J.D., King, V.M., Smith,
Duffield, G.E., Best, J.D., Meurers, B.H., Bittner, A., Loros, J.J., and Dunlap,
A.G., Gant, T.W., Hastings, M.H., and Kyriacou, C.P. (2002). Circadian cycling
J.C. (2002). Circadian programs of transcriptional activation, signaling, and
of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by
protein turnover revealed by microarray analysis of mammalian cells. Curr.
the suprachiasmatic nucleus. Curr. Biol. 12, 540–550.
Biol. 12, 551–557.
Albrecht, U., and Eichele, G. (2003). The mammalian circadian clock. Curr.
Dzhagalov, I., Zhang, N., and He, Y.W. (2004). The roles of orphan nuclear
Opin. Genet. Dev. 13, 271–277.
receptors in the development and function of the immune system. Cell. Mol.
Balsalobre, A., Damiola, F., and Schibler, U. (1998). A serum shock induces cir-
Immunol. 1, 401–407.
cadian gene expression in mammalian tissue culture cells. Cell 93, 929–937.
Eide, E.J., Vielhaber, E.L., Hinz, W.A., and Virshup, D.M. (2002). The circadian
Belden, W.J., Loros, J.J., and Dunlap, J.C. (2007). Execution of the circadian
regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase
negative feedback loop in Neurospora requires the ATP-dependent chroma-
Iepsilon. J. Biol. Chem. 277, 17248–17254.
tin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25, 587–600.
Eide, E.J., Woolf, M.F., Kang, H., Woolf, P., Hurst, W., Camacho, F., Vielhaber,
Bitterman, K.J., Anderson, R.M., Cohen, H.Y., Latorre-Esteves, M., and
E.L., Giovanni, A., and Virshup, D.M. (2005). Control of mammalian circadian
Sinclair, D.A. (2002). Inhibition of silencing and accelerated aging by nicotin-
rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation.
amide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol.
Mol. Cell. Biol. 25, 2795–2807.
Chem. 277, 45099–45107.
Etchegaray, J.P., Lee, C., Wade, P.A., and Reppert, S.M. (2003). Rhythmic
Blander, G., and Guarente, L. (2004). The Sir2 family of protein deacetylases.
histone acetylation underlies transcription in the mammalian circadian clock.
Annu. Rev. Biochem. 73, 417–435.
Nature 421, 177–182.
Blander, G., Olejnik, J., Krzymanska-Olejnik, E., McDonagh, T., Haigis, M.,
Gallego, M., and Virshup, D.M. (2007). Post-translational modifications regu-
Yaffe, M.B., and Guarente, L. (2005). SIRT1 shows no substrate specificity
late the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8, 139–148.
in vitro. J. Biol. Chem. 280, 9780–9785.
Godinho, S.I., Maywood, E.S., Shaw, L., Tucci, V., Barnard, A.R., Busino, L.,
Bordone, L., Motta, M.C., Picard, F., Robinson, A., Jhala, U.S., Apfeld, J.,
Pagano, M., Kendall, R., Quwailid, M.M., Romero, M.R., et al. (2007). The af-
McDonagh, T., Lemieux, M., McBurney, M., Szilvasi, A., et al. (2006). Sirt1 reg-
ter-hours mutant reveals a role for Fbxl3 in determining mammalian circadian
ulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol.
period. Science 316, 897–900.
4, e31. 10.1371/journal.pbio.0040031.
Grozinger, C.M., Chao, E.D., Blackwell, H.E., Moazed, D., and Schreiber, S.L.
Brown, S.A., Ripperger, J., Kadener, S., Fleury-Olela, F., Vilbois, F., Rosbash,
(2001). Identification of a class of small molecule inhibitors of the sirtuin family
M., and Schibler, U. (2005). PERIOD1-associated proteins modulate the neg-
of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276,
ative limb of the mammalian circadian oscillator. Science 308, 693–696.
Brunet, A., Sweeney, L.B., Sturgill, J.F., Chua, K.F., Greer, P.L., Lin, Y., Tran, H.,
Hardin, P.E., Hall, J.C., and Rosbash, M. (1990). Feedback of the Drosophila
Ross, S.E., Mostoslavsky, R., Cohen, H.Y., et al. (2004). Stress-dependent reg-
period gene product on circadian cycling of its messenger RNA levels. Nature
ulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303,
Hirayama, J., Sahar, S., Grimaldi, B., Tamaru, T., Takamatsu, K., Nakahata, Y.,
Busino, L., Bassermann, F., Maiolica, A., Lee, C., Nolan, P.M., Godinho, S.I.,
and Sassone-Corsi, P. (2007). CLOCK-mediated acetylation of BMAL1 con-
Draetta, G.F., and Pagano, M. (2007). SCFFbxl3 controls the oscillation of
trols circadian function. Nature 450, 1086–1090.
Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc. 327
Kaasik, K., and Lee, C.C. (2004). Reciprocal regulation of haem biosynthesis
Ripperger, J.A., and Schibler, U. (2006). Rhythmic CLOCK-BMAL1 binding to
and the circadian clock in mammals. Nature 430, 467–471.
multiple E-box motifs drives circadian Dbp transcription and chromatin transi-
Kornmann, B., Schaad, O., Bujard, H., Takahashi, J.S., and Schibler, U. (2007).
tions. Nat. Genet. 38, 369–374.
System-driven and oscillator-dependent circadian transcription in mice with
Rodgers, J.T., Lerin, C., Haas, W., Gygi, S.P., Spiegelman, B.M., and Puig-
a conditionally active liver clock. PLoS Biol. 5, e34. 10.1371/journal.pbio.
server, P. (2005). Nutrient control of glucose homeostasis through a complex
of PGC-1alpha and SIRT1. Nature 434, 113–118.
Kreppel, F., Biermann, V., Kochanek, S., and Schiedner, G. (2002). A DNA-
Rutter, J., Reick, M., Wu, L.C., and McKnight, S.L. (2001). Regulation of clock
based method to assay total and infectious particle contents and helper virus
and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293,
contamination in high-capacity adenoviral vector preparations. Hum. Gene
Ther. 13, 1151–1156.
Rutter, J., Reick, M., and McKnight, S.L. (2002). Metabolism and the control of
Lavery, D.J., and Schibler, U. (1993). Circadian transcription of the cholesterol
circadian rhythms. Annu. Rev. Biochem. 71, 307–331.
7 alpha hydroxylase gene may involve the liver-enriched bZIP protein DBP.
Sanada, K., Okano, T., and Fukada, Y. (2002). Mitogen-activated protein ki-
Genes Dev. 7, 1871–1884.
nase phosphorylates and negatively regulates basic helix-loop-helix-PAS
Lee, C., Etchegaray, J.P., Cagampang, F.R., Loudon, A.S., and Reppert, S.M.
transcription factor BMAL1. J. Biol. Chem. 277, 267–271.
(2001). Posttranslational mechanisms regulate the mammalian circadian
Sato, T.K., Panda, S., Miraglia, L.J., Reyes, T.M., Rudic, R.D., McNamara, P.,
clock. Cell 107, 855–867.
Naik, K.A., FitzGerald, G.A., Kay, S.A., and Hogenesch, J.B. (2004). A func-
Li, X., Zhang, S., Blander, G., Tse, J.G., Krieger, M., and Guarente, L. (2007).
tional genomics strategy reveals Rora as a component of the mammalian
SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol.
circadian clock. Neuron 43, 527–537.
Cell 28, 91–106.
Shirogane, T., Jin, J., Ang, X.L., and Harper, J.W. (2005). SCFbeta-TRCP con-
Liu, C., Li, S., Liu, T., Borjigin, J., and Lin, J.D. (2007). Transcriptional coactiva-
trols clock-dependent transcription via casein kinase 1-dependent degrada-
tor PGC-1alpha integrates the mammalian clock and energy metabolism.
tion of the mammalian period-1 (Per1) protein. J. Biol. Chem. 280, 26863–
Nature 447, 477–481.
Lowrey, P.L., and Takahashi, J.S. (2000). Genetics of the mammalian circadian
Siepka, S.M., Yoo, S.H., Park, J., Song, W., Kumar, V., Hu, Y., Lee, C., and
system: Photic entrainment, circadian pacemaker mechanisms, and post-
Takahashi, J.S. (2007). Circadian mutant Overtime reveals F-box protein
translational regulation. Annu. Rev. Genet. 34, 533–562.
FBXL3 regulation of cryptochrome and period gene expression. Cell 129,
McBurney, M.W., Yang, X., Jardine, K., Hixon, M., Boekelheide, K., Webb,
J.R., Lansdorp, P.M., and Lemieux, M. (2003). The mammalian SIR2alpha
Stokkan, K.A., Yamazaki, S., Tei, H., Sakaki, Y., and Menaker, M. (2001).
protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23,
Entrainment of the circadian clock in the liver by feeding. Science 291, 490–
Motta, M.C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma,
Storch, K.F., Lipan, O., Leykin, I., Viswanathan, N., Davis, F.C., Wong, W.H.,
Y., McBurney, M., and Guarente, L. (2004). Mammalian SIRT1 represses fork-
and Weitz, C.J. (2002). Extensive and divergent circadian gene expression in
head transcription factors. Cell 116, 551–563.
liver and heart. Nature 417, 78–83.
Nagoshi, E., Saini, C., Bauer, C., Laroche, T., Naef, F., and Schibler, U. (2004).
Takata, T., and Ishikawa, F. (2003). Human Sir2-related protein SIRT1 associ-
Circadian gene expression in individual fibroblasts: cell-autonomous and self-
ates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and
sustained oscillators pass time to daughter cells. Cell 119, 693–705.
HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun.
Naruse, Y., Oh-hashi, K., Iijima, N., Naruse, M., Yoshioka, H., and Tanaka, M.
(2004). Circadian and light-induced transcription of clock gene Per1 depends
Tu, B.P., and McKnight, S.L. (2006). Metabolic cycles as an underlying basis of
on histone acetylation and deacetylation. Mol. Cell. Biol. 24, 6278–6287.
biological oscillations. Nat. Rev. Mol. Cell Biol. 7, 696–701.
Panda, S., Antoch, M.P., Miller, B.H., Su, A.I., Schook, A.B., Straume, M.,
Vaziri, H., Dessain, S.K., Ng Eaton, E., Imai, S.I., Frye, R.A., Pandita, T.K.,
Schultz, P.G., Kay, S.A., Takahashi, J.S., and Hogenesch, J.B. (2002). Coordi-
Guarente, L., and Weinberg, R.A. (2001). hSIR2(SIRT1) functions as an NAD-
nated transcription of key pathways in the mouse by the circadian clock. Cell
dependent p53 deacetylase. Cell 107, 149–159.
Walker, J.R., and Hogenesch, J.B. (2005). RNA profiling in circadian biology.
Preitner, N., Damiola, F., Lopez-Molina, L., Zakany, J., Duboule, D., Albrecht,
Methods Enzymol. 393, 366–376.
U., and Schibler, U. (2002). The orphan nuclear receptor REV-ERBalpha con-
Wallberg, A.E., Yamamura, S., Malik, S., Spiegelman, B.M., and Roeder, R.G.
trols circadian transcription within the positive limb of the mammalian circa-
(2003). Coordination of p300-mediated chromatin remodeling and TRAP/
dian oscillator. Cell 110, 251–260.
mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149.
Preitner, N., Brown, S., Ripperger, J., Le-Minh, N., Damiola, F., and Schibler,
Yagita, K., Tamanini, F., van Der Horst, G.T., and Okamura, H. (2001). Molec-
U. (2003). Orphan nuclear receptors, molecular clockwork, and the entrain-
ular mechanisms of the biological clock in cultured fibroblasts. Science 292,
ment of peripheral oscillators. Novartis Found. Symp. 253, 89–99.
Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E.,
Yagita, K., Tamanini, F., Yasuda, M., Hoeijmakers, J.H., van der Horst, G.T.,
Wilm, M., and Seraphin, B. (2001). The tandem affinity purification (TAP)
and Okamura, H. (2002). Nucleocytoplasmic shuttling and mCRY-dependent
method: a general procedure of protein complex purification. Methods 24,
inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21, 1301–
Reick, M., Garcia, J.A., Dudley, C., and McKnight, S.L. (2001). NPAS2: an an-
Yoo, S.H., Ko, C.H., Lowrey, P.L., Buhr, E.D., Song, E.J., Chang, S., Yoo, O.J.,
alog of clock operative in the mammalian forebrain. Science 293, 506–509.
Yamazaki, S., Lee, C., and Takahashi, J.S. (2005). A noncanonical E-box en-
Reppert, S.M., and Weaver, D.R. (2002). Coordination of circadian timing in
hancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad.
mammals. Nature 418, 935–941.
Sci. USA 102, 2608–2613.
328 Cell 134, 317–328, July 25, 2008 ª2008 Elsevier Inc.
Source: http://www.molbio.unige.ch/document/name/18662546.pdf
Acute Otitis Media: For the treatment of acute otitis media in pediatric patients due to suscep- tible strains of Streptococcus pneumoniae or Haemophilus influenzae when in the judgment ofthe physician sulfamethoxazole and trimethoprim offers some advantage over the use of other Susceptibility Testing Methods: antimicrobial agents. To date, there are limited data on the safety of repeated use of BACTRIM
ARTICLESPUBLISHED ONLINE: 23 NOVEMBER 2008 DOI: 10.1038/NPHYS1148 Multiphase transformation and Ostwald's rule ofstages during crystallization of a metal phosphate Sung-Yoon Chung1,2*, Young-Min Kim3, Jin-Gyu Kim3 and Youn-Joong Kim3 Although the classical picture of crystallization depicts a simple and immediate transformation from an amorphous to








