458 isolation

Full Length Article
Amit Pandey* , Neha Rajput , Mohammad Nadeem Aslam and Bharat Choudhary
1. 1R&D Division, MRD LifeSciences, Lucknow, India. 2. Alpine Institute of Management and Technology, Dehradun, India. 3. APS University, Rewa, M.P., India
Five MDR cultures of bacteria were isolated from four different locations of Lucknow and Kanpur, respectively. Based upon Gram staining four isolates (A1, A2, C2 and G3) were identified as Gram negative out of which A1 was mixed culture that is cocci + rod and A2 and G3 were Gram negative rods. Culture N was identified as Gram positive mixed culture that is cocci + rod. Based upon the biochemical analysis G3 bacterial culture was identified as Pseudomonas aeruginosa (Table 1), for multi-drug resistance (MDR) test seven different antibiotics in different concentration were used. It was observed that A1 bacterial culture showed resistance to amoxycilline (100 µg/ml), cefixime (100 µg/ml), ofloxacin (50 µg/ml), roxithromycin (20 µg/ml), ampicilline (50 µg/ml) and tetracycline (50 µg/ml) while A2 culture was resistant to amoxycilline (50 µg/ml) and cefixime (1 mg/ml). C2 was resistant to amoxycilline (10 mg/ml), cefixime (1 mg/ml), roxithromycin (10 mg/ml), ampicilline (20 µg/ml) and tetracycline (20 µg/ml). G3 was resistant to amoxycillin (1 mg/ml) and cefixime (1 mg/ml). And N culture was resistant to amoxycilline (100 µg/ml), cefixime (1 mg/ml), roxithromycin (50 µg/ml), ampicilline (50 µg/ml) and tetracycline (20 µg/ml).
Keywords: Multi-Drug Resistance (MDR), Susceptibility; Antibiotics; Bacterial Isolates; Broad Spectrum; Minimum
Inhibitory Concentration (MIC).
Abbreviations : µg- Microgram; ml- Mililitre; mg - Miligram; R - Resistant; S- Sensitive; AR-Antibiotic Resistance;
hrs.-Hours; MBC -Minimum Bactericidal Concentration; MIC- Minimum Inhibitory Concentration;
MDR- Multi-Drug Resistance
bacterium to be effective against MDR bacterial cultures. The
microorganisms have a defense system against various antibiotics and
Antibiotic resistance is a considerable term while studying about
they grow even in presence of antibiotics by getting resistant. It was
diseases in human, animals and plants. Mostly the antibiotic resistant
suggested that soil microbes harbour antibiotic resistant (AR) genes with
bacterial cultures are found in clinical and veterinary land areas whereas
a diverse gene sequencing (Jalal, et al., 2010). The antibiotic sensitivity
the existence of antibiotic resistance (AR) bacteria in water has also
is expressed in terms of minimal inhibitory concentration (MIC) and
observed and supported to be present in higher concentrations and
minimal bactericidal concentration (MBC) and it give quantitative data.
diversity in hospital areas as compared to domestic areas. Antibiotics are
These quantitative results are useful in predicting the tissue, blood or
either synthesized industrially or also produced by microorganisms;
urine levels of antibiotics that must be attained to assure inhibition or
these antibiotics have microstatic or microcidal activity. Mainly these
killing. In comparison to clinical or hospital area soil, the soil from
antibiotics producing microbes interrupt the microbial metabolism by
various aquatic places were taken for isolating the AR pathogens. The
various mechanisms (Jalal et al., 2010). Several researches were
main objective of this work is to isolate the multi--drug resistant (MDR)
performed that dealt with advanced antimicrobial resistance in bacteria
pathogens from the aquatic places of Lucknow and Kanpur, U.P against
isolated from food, animal and environment (Jensen et al., 2001). The
various antibiotics and to know the MIC and MBC values for various
use of antibiotics is increasing continuously in different fields like
antibiotics in presence of the isolated pathogens.
veterinary medicine, agriculture, medicines etc. but the awareness of knowledge regarding the quantity of antibiotics present in the
Materials and Methods
environment after their use is very less (Hirsch et al., 1998). The population of antibiotic resistance (AR) bacteria in the areas where
The soil samples were collected from five different locations of
antibiotics are used is common but existence of AR bacteria in aquatic
Lucknow and Kanpur; A (Sewage water, Lucknow), C (Pond water,
environment is also increasing readily (Schwartz et al., 2003). It is
Lucknow), G (Ganga river, Kanpur) and N (Gomti river, Lucknow).
observed that resistance in bacteria could be – (i) intrinsic resistance i.e.
Samples were serially diluted and bacteria were isolated on Nutrient
natural resistance of bacteria to certain antibiotics. (ii) Acquired resistance i.e. the susceptible that become resistant by adapting itself
through genetic changes. (iii) Multi--drug resistance i.e. the resistance of
E- Mail: [email protected]
Full Length Article
Isolation and Characterization of Multi-Drug Resistant Cultures, Amit Pandey et al.,
Biochemical tests
Isolate C2 Isolate G3
Agar, two bacterial colonies were selected and designated as: A1, A2, C1, C2, G1, G2, N1 and
N2, respectively. Bacterial colonies were sub
Cellular morphology
cultured several times up to pure culture,
Catalase activity
maintained for further biochemical analysis,
and preserved at low temperature. After isolation, identification of the bacterial isolates
Voges proskauer's test
was carried out according to Bergey's Manual of
Citrate utilization
Determinative Bacteriology (Buchanan and
Gibbonns, 1978).
All the bacterial isolates were analyzed for their
King's B medium test
sensitivity against commonly used antibiotics
(Pseudomonas
by agar well diffusion method. Different
antibiotics having different mechanisms of action were taken as tetracycline, amoxycilline,
ampicilline, roxithromycin, ofloxacin, cefixime Table 1. Biochemical analysis of isolated MDR bacterial cultures.
and ciprofloxacin of concentration 5 µg/ml, 10 µg/ml, 20 µg/ml, 30 µg/ml, 50 µg/ml, 100
µg/ml, 1 mg/ml and 10 mg/ml. Wells were
prepared on nutrient agar plate previously
spreaded with 50 µl of isolated bacterial broth
culture. These wells were loaded with 50µl of
antibiotic solution and then plates were
incubated at 37°C for overnight, then the zone of
inhibition were examined. The obtained multi-
characterized by microscopic and Biochemical
characterized by staining (Gram staining and
Endospore staining) and biochemical activity (IMVIC) as par Bergey's manual given in Aneja
Table 2. MDR test for sample A1 [Note: R= resistant, S= sensitive].
(2003). King's B medium is a confirmation medium for detection and differentiation of P s e u d o m o n a s a e r u g i n o s a f r o m o t h e r
Pseudomonas based on fluorescein (pyoverdin)
production and pyocyanin inhibition. Peptone,
potassium hydrogen phosphate, heptahydrated
magnesium sulfate (MgSO .7H O) and
bacteriological agar were added to distilled water (pH 7.0 – 7.2). Solution was autoclaved and
poured in test tube to prepare slants. The isolates
were inoculated on slants and kept for incubation
at 37°C for 48 hrs.
MIC is the lowest concentration which resulted
in maintenance or reduction of inoculum viability". Only antibiotic that shows inhibitory
Table 3. MDR test for sample A2 [Note: R= resistant, S= sensitive].
activity toward the bacterial isolate using the Kirby-Bauer methods are tested further. The active antibiotics were serially diluted to make a range of antibiotic
concentrations that encompasses the concentration used in the (Kirby-
Five MDR cultures of bacteria were isolated from four different
Bauer method, 1966). 20 µl of test bacterium was added to all tubes
locations of Lucknow and Kanpur. Based upon Gram staining, four
having 3 ml of nutrient broth which was serially diluted with antibiotic.
isolates (A1, A2, C2 and G3) were identified as Gram negative out of
After incubation, the MIC is identified as the least concentration of
which A1 was mixed culture i.e. cocci + rod and A2 and G3 were Gram
antibiotic that inhibits the growth of the test bacterium. The Minimal
negative rods. Culture N was identified as Gram positive mixed culture
Bactericidal Concentration (MBC) is determined by spreading the
that is cocci + rod. All isolated bacterial cultures were aerobic. Based
bacterial colonies on nutrient agar plate which was obtained by MIC.
upon the biochemical analysis, G3 bacterial culture was identified as Pseudomonas aeruginosa (Table 1). For the multi- drug resistance
Full Length Article
Authors to provide, P.A. Mary Helen et al.,
Isolation and Characterization of Multi-Drug Resistant Cultures, Amit Pandey et al.,
Concentration Amoxy
(MDR) test seven different antibiotics in different
concentration were used. It was observed that A1
bacterial culture showed resistance to amoxicilline
(100 µg/ml), cefixime (100 µg/ml), ofloxacin (50
µg/ml), roxithromycin (20 µg/ml), ampicilline (50
µg/ml) and tetracycline (50 µg/ml). A2 culture was
resistant to amoxicilline (50 µg/ml) and cefixime (1
mg/ml). C2 was resistant to amoxicilline (10
mg/ml), cefixime (1 mg/ml), roxithromycin (10
mg/ml), ampicillin (20 µg/ml) and tetracycline (20
µg/ml). G3 was resistant to amoxicilline (1mg/ml)
and cefixime (1 mg/ml). And N culture was
Table 4. MDR test for sample C2 [Note: R= resistant, S= sensitive].
resistant to amoxicilline (100 µg/ml), cefixime (1 mg/ml), roxithromycin (50 µg/ml), ampicilline (50 µg/ml) and tetracycline (20 µg/ml). It was observed
Concentration Amoxy
that even after showing resistance at higher
concentrations amoxicilline was sensitive at
concentration 20 µg/ml against four cultures (A2, C2, G3 and N) which was considered as MIC value
of amoxicilline. Roxithromycin also showed
sensitivity at concentration 30 µg/ml against
culture N which was considered as its MIC value.
Antibiotics are chemotherapeutic agents that have
revolutionized the treatment of infectious disease – turning life-threatening diseases into more
Table 5. MDR test for sample G3 [Note: R= resistant, S= sensitive].
manageable and treatable conditions. Resistance became a major challenge to the treatment of
Concentration Amoxy
infectious diseases shortly after the introduction of
antibiotics. The objective of the experiment was to
isolate multi-drug resistant bacteria which can use antibiotics as carbon source for their nutrition and
growth. Bacteria gain resistance through various
methods: some bacteria make an antibiotic
ineffective before the drug can kill them; some
strains alter the drug attack site so that the
antibiotic becomes ineffective; some rapidly pump
out the antibiotic–antibiotic efflux. Some bacteria
have a natural resistance to antibiotics but others
become resistant through genetic mutation or by
acquiring resistance from another bacterium
Table 6. MDR test for sample N [Note: R= resistant, S= sensitive].
(Alliance for the Prudent Use of Antibiotics, 1999). Five multi-drug resistant bacterial cultures were isolated from all sampling points. Out of which only one was identified as Pseudomonas
aeruginosa. This culture causes infections of wounds, burns, eyes and
ears (Lateef, 2003). The antibiotic sensitivity pattern of the five bacterial
isolates is shown in Tables (2–6). All five cultures were resistant to
cefixime and amoxycillin even at higher concentrations (100 µg), which was much higher compare to work done by Lateef (2003) this process
was done with the help of Agar well diffusion method which was earlier
done by Bauer et al. (1966). Only A1 isolate was resistant to
ofloxacin.A1, C2 & N isolates were resistant to roxithromycin, ampicillin and tetracycline. None of the five isolates were resistant to
Table 7. MIC value of culture A2 against Cefixime
ciprofloxacin i.e. all were susceptible to ciprofloxacin. After MIC the
(Initial concentration= 1mg/ml)
antibiotics amoxycillin, cefixime showed resistance at higher concentration (0.25 mg/ml). After MBC it was observed that the MDR


Full Length Article
Isolation and Characterization of Multi-Drug Resistant Cultures, Amit Pandey et al.,
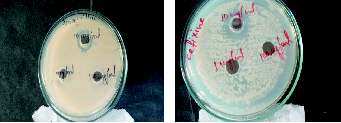
Figure 1. Resistance shown by isolated MDR culture C at higher
Table 8. MIC value of culture C2 against Cefixime
concentration (100 µg/ml, 1 mg/ml & 10 mg/ml) of cefixime
(Initial concentration= 1mg/ml)
and amoxycilline
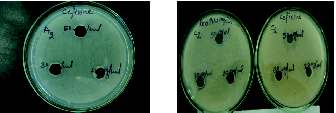
Figure 2. Resistance shown by MDR isolates at lower concentrations of cefixime and roxithromycin for cultures A2 & C2
Table 9. MIC value of culture C2 against Amoxycilline (Initial concentration= 1mg/ml)
Figure 3. Resistance shown by isolates N & A2 with amoxycilline
Table 10. MIC value of culture A2 against Amoxycilline (Initial concentration= 1mg/ml)
cultures are bacteriostatic as they show growth even at higher concentrations of antibiotics.
At the end of all the experiments it was identified that out of all bacterial cultures isolated from various aquatic places, five were multi--drug
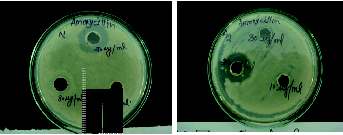
Figure 4. MBC culture plates of the isolates (Cultures showing the
resistant. Isolates A2 & C2 are Gram negative rods, isolates A1 & G3 are
growth in the presence of Antibiotics) A2: culture against Cefixime Gram
negative mixed cultures and only one isolate N is Gram positive
C2: culture against Amoxycilline
mixed culture, all isolates were catalase positive. Isolate G3 was identified as Pseudomonas aeruginosa and A2 of family Enterobacteriaceae. All these five isolates were multi--drug resistant as they were resistant to various antibiotics used which were of different mechanism of action. After MIC & MBC tests it was observed that the isolated cultures were bacteriostatic in nature.
I am very grateful and my heartiest thanks to Mr. Manoj Verma, Director, MRD LifeSciences (P) Limited, Lucknow, Mr. R.P. Mishra (Research Scientist), Mr. Jahir Alam Khan (Research Scientist), & Ms. Chanda Sinha (Research Scientist), MRDLS, Lucknow, for there kind support
Figure 5. Confirmatory test of Pseudomonas aeruginosa in G3 isolate
throughout the research work. I am also thankful to the almighty without
(Pink colonies in King's B Media).
Full Length Article
Isolation and Characterization of Multi-Drug Resistant Cultures, Amit Pandey et al.,
whose consent anything is possible.
Flavobacterium/Bacteroides line of phylogenetic descent. Syst. Appl. Microbiol. 21: 374-383.
Jalal, K.C.A., Nur Fatin U.T., Mardiana M.A., Akbar John B., Kamaruzzaman Y.B., et al., 2010. Antibiotic resistance microbes in
Aneja, K.R., 2003. Experiments in Microbiology, Plant Pathology and
tropical mangrove sediments in east coast peninsular, Malaysia. African
Biotechnology. New Age International (P) Limited: New Delhi Vol. 4. P.
Journal of Microbiology Research. 8:640-645
Jensen LB., Baloda S., Boye M., Aarestrup FM. 2001. Antimicrobial
Alliance for the Prudent Use of Antibiotics, 1999. Antimicrobials in the
resistance among Pseudomonas spp. and the Bacillus cereus group
United States. Strategies Developed by an NGO: 1-5.
isolated from Danish agricultural soil. Environ. Int. 26: 581-587
Bauer AW., Kirby WM., Sheris J.C., Turck M. 1966. Antibiotic
Lateef, A., 2003. The microbiology of a pharmaceutical effluent and its
susceptibility testing by a standardized single disc method. Am. J. Clin.
public health implications 3: 212-218.
Pathol. 45: 149-158.
Schwartz T., Kohenen W., Jansen B., Obst U., 2003. Detection of
Buchanan R.E. and Gibbonns E.N. 1978. Bergey's Manual of
antibiotic resistant bacteria and their resistance genes in waste water,
Determinative Bacteriology. Williams and Wilken Co.: Baltimore surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 43:
Hirsch P., Ludwig W., Hethke C., Sittig M., Hoffmann B., Gallikowski
Vincent D.S.J. and DVM., MA., 1999 Informing Public Policy on
CA., 1998. Hymenobacter roseosalivarius gen. Nov. From continental
Agricultural Use of
Antarctic soils and sandstone: bacteria of the Cytophaga/
Citation: Amit P., Neha R., Mohammad N A., Bharat C. 2011. Isolation and characterization of multi-drug resistant cultures from aquatic areas. Adv Bio Tech 11(4):16-20
Source: http://www.mrdlifesciences.com/admin/force_download.php?action=rnp&f=Latest%20Research%20Paper@MRD%20LifeSciences-19.pdf1320232737.pdf
PHILIPPINE BIDDING DOCUMENTS (As Harmonized with Development Partners) PROCUREMENT OF MEDICINES AND MEDICAL & DENTAL SUPPLIES & EQUIPMENT (REBID) BFP BAC IB No. 2013-05(R) Government of the Republic of the Philippines BUREAU OF FIRE PROTECTION National Headquarters Fourth Edition These Philippine Bidding Documents (PBDs) for the procurement of Goods through
Update of Pharmacological Intervention Recommendations for the Canadian Consensus Conference on the Diagnosis and Treatment of Dementia 2012 1) New recommendation for the management of Alzheimer's disease New Recommendation: Many cases of dementia have more than one condition contributing to causation. Most commonly this will be a combination of Alzheimer's disease with other brain pathology. It is recommended that management be based on what is (are) felt to be the predominant contributing cause(s). (Grade 1B)







