Dudley 1.6
Computational Repositioning of the AnticonvulsantTopiramate for Inflammatory Bowel Disease
Joel T. Dudley,1,2,3* Marina Sirota,1,2,3* Mohan Shenoy,4 Reetesh K. Pai,5Silke Roedder,1,3 Annie P. Chiang,1,2,3 Alex A. Morgan,1,2,3 Minnie M. Sarwal,1,3Pankaj Jay Pasricha,4 Atul J. Butte1,3†
Inflammatory bowel disease (IBD) is a chronic inflammatory disorder of the gastrointestinal tract for which thereare few safe and effective therapeutic options for long-term treatment and disease maintenance. Here, we ap-plied a computational approach to discover new drug therapies for IBD in silico, using publicly available moleculardata reporting gene expression in IBD samples and 164 small-molecule drug compounds. Among the topcompounds predicted to be therapeutic for IBD by our approach were prednisolone, a corticosteroid used to treatIBD, and topiramate, an anticonvulsant drug not previously described to have efficacy for IBD or any related dis-
orders of inflammation or the gastrointestinal tract. Using a trinitrobenzenesulfonic acid (TNBS)
–induced rodentmodel of IBD, we experimentally validated our topiramate prediction in vivo. Oral administration of topiramatesignificantly reduced gross pathological signs and microscopic damage in primary affected colon tissue in theTNBS-induced rodent model of IBD. These findings suggest that topiramate might serve as a therapeutic optionfor IBD in humans and support the use of public molecular data and computational approaches to discover newtherapeutic options for disease.
compendium of gene expression signatures comprising 164 drug
Inflammatory bowel disease (IBD), of which Crohn's disease (CD)
compounds to infer previously undescribed therapeutic relationships
and ulcerative colitis (UC) are the most common clinically defined
between drug-disease pairs represented in the data sets. Among the
manifestations, represents a group of chronic, progressive inflamma-
highest-scoring therapies predicted from our approach was the cor-
tory disorders of the intestinal tract that affects more than 1 million
ticosteroid prednisolone, a known treatment for IBD. The approved
individuals in North America alone (1). There is currently no known
antiepileptic drug topiramate, which has not previously been de-
cure for IBD, and available treatment options are aimed toward con-
scribed to have a therapeutic association with IBD or any other in-
trolling symptoms, promoting remission, and preventing relapse. Cur-
testinal disorders, was unexpectedly ranked higher by our method
rent treatment protocols for IBD incorporate chronic administration of
(that is, higher predicted therapeutic score for IBD) than predniso-
corticosteroids and systemic anti-inflammatory drugs to reduce or inhib-
lone. We therefore evaluated the efficacy of topiramate for IBD in
it primary inflammation, along with antibiotics to treat secondary in-
three independent experiments using a trinitrobenzenesulfonic acid
fection (2). Chronic use of corticosteroids or systemic anti-inflammatory
(TNBS)–induced rodent model of colitis.
drugs (for example, 6-mercaptopurine) typically used to treat IBD isassociated with severe side effects, and more targeted therapies such asanti-TNFa (tumor necrosis factor–a) drugs incur high costs and elicit
a therapeutic response in only a subset of affected patients (3). Surgicalremoval of affected regions of the small or large intestine is also used
Prediction of known and previously undescribed
as a treatment strategy. This option is expensive and highly invasive,
and the disease can often remanifest in previously unaffected locations
To discover new therapeutic agents for IBD, we compared the gene
along the intestinal tract (2).
expression profiles from a compendium of 164 drug compounds (5)
Here, we aimed to discover and validate new therapeutic options
to a gene expression signature of IBD derived from publicly available
for IBD, using a systematic computational approach for drug reposi-
experiments obtained from National Center for Biotechnology Infor-
tioning that is based on integration of public gene expression signatures
mation (NCBI) Gene Expression Omnibus (GEO) (6). We derived
of drugs and diseases (4). We systematically evaluated gene expres-
therapeutic predictions for drug-disease pairs based on the hypothesis
sion signatures of IBD derived from public microarray data against a
that if a drug has a gene expression signature that is opposite of adisease signature, that drug could potentially be used as a treatmentfor that disease. Our method produces a negative score when the drug
1Division of Systems Medicine, Department of Pediatrics, Stanford University School of
signature is oppositional to the disease signature and a positive score
Medicine, 251 Campus Drive, Stanford, CA 94305–5415, USA. 2Training Program in
when they are concordant (see Materials and Methods). Among the
Biomedical Informatics, Stanford University School of Medicine, Stanford, CA 94305–5479, USA. 3Lucile Packard Children's Hospital, 725 Welch Road, Palo Alto, CA 94304, USA.
strongest therapeutic predictions for CD is the corticosteroid prednis-
4Division of Gastroenterology, Department of Medicine, Stanford University School of
olone (score = −0.216), a well-known treatment for these conditions
Medicine, 300 Pasteur Drive, Stanford, CA 94305–5187, USA. 5Department of Pathology,
(7) (Fig. 1). We observed that topiramate, an anticonvulsant drug cur-
Stanford University School of Medicine, Stanford, CA 94305–5324, USA.
rently used to treat epilepsy, had a stronger therapeutic score for
*These authors contributed equally to this work.
†To whom correspondence should be addressed. E-mail:
[email protected]
Crohn's (−0.220) (Fig. 1, red arrow) than the established therapeutic
Vol 3 Issue 96 96ra76
Drug-disease score
prednisolone. Based on our analysis, topiramate is also one of thestrongest predicted therapies for UC, with a score of
−0.219. Although
another compound, an isoindoline carboxamide (ChemBridge 5186324),
scored higher than topiramate by our method, we focused on topiramate
for experimental validation because it is a Food and Drug Administra-
tion (FDA)–approved compound known to be generally safe in humans
and is readily available for clinical use.
Experimental validation of topiramate as an
indication for IBD
To determine whether our in silico drug indication predictions would
translate into therapeutic efficacy in vivo, we tested whether topiramate
15-Delta prostaglandin J2
would show efficacy for IBD by means of a TNBS-induced rat model
of IBD. We performed an initial pilot validation experiment followed
by two independent replication experiments in male Sprague-Dawley
rats given TNBS [5% (w/v) total of 100 mg/kg] intrarectally to induce
colitis. One group was not induced with TNBS and was treated with
vehicle only, another group was induced with TNBS and treated with
vehicle (TNBS + vehicle), and the third group was induced with TNBS
and treated with topiramate (80 mg/kg per day) by oral gavage (TNBS
+ topiramate). A fourth group that was induced with TNBS and treated
with prednisolone (3 mg/kg per day) served as a positive control
(TNBS + prednisolone). After the initial TNBS induction, animals
were treated for 7 consecutive days, and the induced IBD phenotype
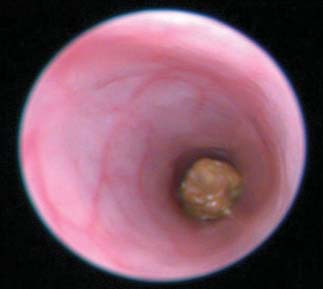
was assessed in vivo by video endoscopy at days 3 and 7.
Disease severity was assessed over the course of treatment by ob-
servation of clinical signs and by gross inspection of affected colon tis-
sues after treatment termination. Animals treated with both prednisolone
(TNBS + prednisolone) and topiramate (TNBS + topiramate) exhibited
reduced incidence of diarrhea over the course of treatment compared
to affected animals administered vehicle alone (TNBS + vehicle) (Fig. 2).
% Animals w/ diarrhea
Tyrphostin AG-825
Fig. 1. Significant drug-disease scores for Crohn's disease (CD). The names of
TNBS + prednisolone
the drugs are placed along the bottom axis, and the vertical bars above the
TNBS + topiramate
drug name indicate the computationally predicted therapeutic score for thedrug based on comparison of the gene expression signature of the drug with
Fig. 2. Effect of topiramate on clinical evaluation of IBD severity. For
the gene expression signature of CD. A positive score indicates that the drug
each study day, treatment groups were scored by percent of animals
exhibits an expression pattern that is synergistic with the disease, whereas a
with diarrhea within each group (n = 12 animals per group). The pre-
negative score indicates that the drug exhibits an expression pattern that is
ponderance of diarrhea was found to be significantly different among
oppositional to the disease. Drugs are sorted from left to right starting with
treatment groups (one-way ANOVA, P < 0.005). IBD-induced animals
those predicted to be most efficacious for the disease. Green bars indicate
treated with topiramate (TNBS + topiramate) exhibited significantly re-
drugs that are discussed in the text. The red triangle points toward the anti-
duced diarrhea over the course of the study compared to the respective
convulsant drug topiramate, which was selected for experimental validation.
control (TNBS + vehicle; Tukey-Kramer test, P < 0.05).
Vol 3 Issue 96 96ra76
Expression signature evaluationThe predicted efficacious relationship be-tween IBD and topiramate was inferredfrom public data, suggesting that partic-ular sets of genes would exhibit oppo-sitional expression between drug anddisease. Comparative visual inspection ofthe Crohn's and topiramate expression
Vehicle only
TNBS + vehicle
Vehicle only
TNBS + vehicle
signatures revealed the expected antithet-ical expression patterns between the drugand the disease, and functional enrichmentanalysis indicated that genes involved withgastrointestinal disease, inflammatory re-sponse, and other immune-related functionswere reciprocally expressed between thedrug-affected and the disease-affected con-
TNBS + topiramate
TNBS + prednisolone
TNBS + topiramate
TNBS + prednisolone
ditions (Fig. 4A).
We performed quantitative polymer-
ase chain reaction (qPCR) analysis spotchecks of postmortem colon tissues to eval-
uate whether the expected expression pat-
terns of genes reflected in the public gene
expression data driving our prediction
were observed in the animal validation
study. We randomly selected eight genes
Gross pathology score
Microscopic damage score
for qPCR analysis from those genes ex-hibiting opposing expression patterns
between drug and disease expression sig-
TNBS + veh
TNBS + top
TNBS + veh
TNBS + top
TNBS + pred
TNBS + pred
natures and for which commercial pri-mers were available (see Materials and
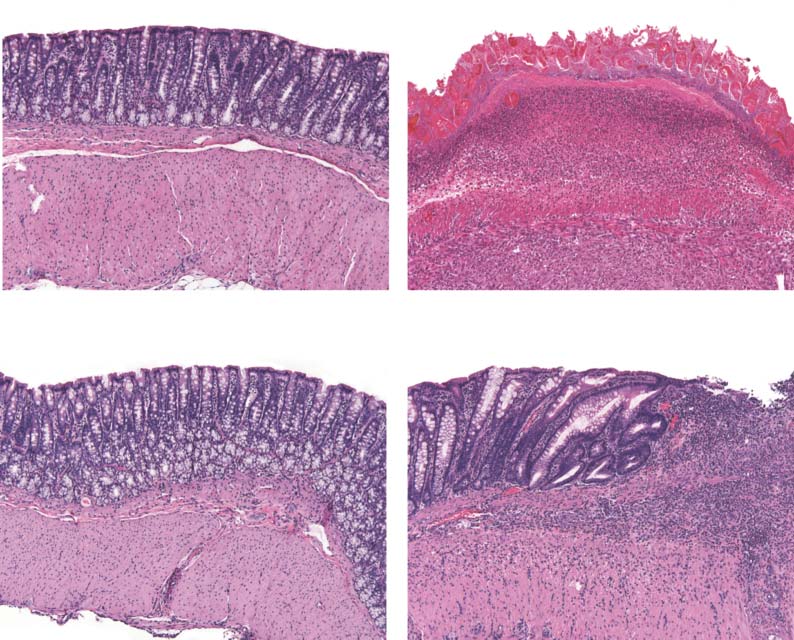
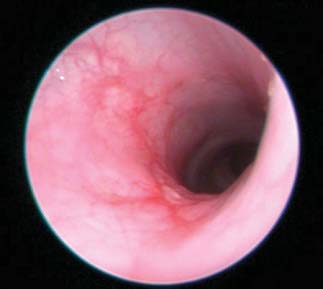
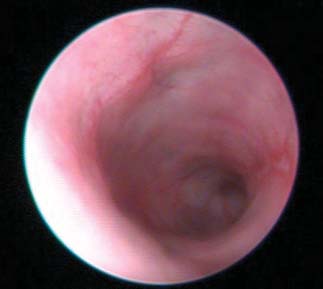
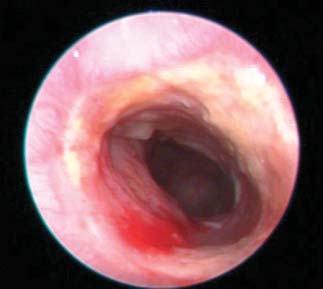
Fig. 3. Effect of topiramate on pathological assessment of IBD severity. (A) Clinical endoscopy capturedfrom live animals on day 7 of the study. (B) Gross pathology score. (C) Micrographs of H&E-stained colon
Methods). qPCR analysis revealed that
tissues showing microscopic damage to the mucosal and epithelial layers of the colon wall between treat-
two of these genes, TRPV1 and IFI30, were
ment groups. (D) Macroscopic damage score assessed from light microscopy of fixed colon tissues. Data
differentially expressed between treat-
graphs represent the mean and SEM estimated from three independent experiments (n = 12 rats per
ment groups in the direction expected
group). *P < 0.05; ****P < 0.00005, two-sided Mann-Whitney U test.
from comparison of the public moleculardata (Fig. 4B). TRPV1 was significantly up-
Assessment of colitis by visual inspection of endoscopy video captured
regulated in the topiramate-treated group (TNBS + topiramate) relative
on day 7 of treatment and scoring of disease severity demonstrated re-
to the untreated disease-induced group (TNBS + vehicle) (P < 0.05,
duced gross pathological inflammation and ulceration in the topiramate-
Mann-Whitney U test; n = 12 per group), and IFI30 was significantly
and prednisolone-treated groups relative to the untreated (TNBS + vehicle)
down-regulated in the topiramate-treated group relative to the un-
group (Fig. 3A and videos S1 to S4). Quantitative assessment of the
treated disease-affected group (TNBS + vehicle) (P < 0.005, Mann-
gross pathological characteristics revealed that animals in the topiramate-
Whitney U test; n = 12 per group). These findings corroborate the
treated group (TNBS + topiramate) exhibited significantly reduced
expected oppositional relationships between these genes reflected in
swelling, ulceration, and other gross pathological characteristics com-
the expression signatures that were used to computationally predict
pared to animals in the untreated (TNBS + vehicle) group (P < 0.0001,
an efficacious relationship between topiramate and IBD.
Mann-Whitney U test; n = 12 per group) (Fig. 3B).
Microscopic damage was assessed by histopathology analysis of
fixed colon tissue sections harvested at the conclusion of the dosing
schedule. Visual inspection of fixed colon tissues revealed extensivedestruction of the colon mucosal layer in the untreated colitis-induced
Using an in silico approach based on the integration of publicly avail-
group (TNBS + vehicle), which was substantially ameliorated in colitis-
able gene expression data, we inferred that the anticonvulsant topiramate
induced animals receiving topiramate (TNBS + topiramate) (Fig. 3C).
was a potential new therapeutic agent for IBD and performed an ex-
Quantitative evaluation demonstrated significantly reduced microscopic
perimental validation that confirmed topiramate's efficacy in ameliorat-
damage in animals treated with topiramate compared to animals in the
ing a TNBS-induced rodent model of IBD. The precise mechanism of
untreated (TNBS + vehicle) group (P < 0.05, Mann-Whitney U test; n = 12
action for topiramate is unknown, but it is known to enhance the ac-
per group) (Fig. 3D). Together, these data provide evidence that topiramate
tivity of g-aminobutyric acid (GABA)–activated chloride channels, ac-
exhibits efficacy against IBD in the TNBS model, as predicted by our
tivate kainate and AMPA receptors, and inhibit the activity of some
carbonic anhydrase enzymes (8). Topiramate is administrated orally
Vol 3 Issue 96 96ra76
- Actin signaling
improvements in both gross and microscopic measures of disease
- Trombin signaling
pathology relative to the relevant vehicle-treated group (TNBS + vehicle),
- Gastrointestinal
with endpoints exceeding even the prednisolone-treated positive con-
trol group. Both topiramate and prednisolone reduced the severity of
- NF-κB signaling
- IL-15 signaling
diarrhea over the course of treatment relative to the vehicle-treated
Crohn's disease signature
Fold change
- B cell receptor
group (TNBS + vehicle); however, only the topiramate-treated group
exhibited significantly reduced gross pathology and microscopic
Univ RNA normalized to 18
damage scores (Fig. 3, B and D). Although prednisolone is an estab-
lished treatment for IBD in humans and was correctly identified as
TNBS + topiramate
therapeutic by our computational method, previous studies have
reported limited efficacy of prednisolone in chemically induced ro-
dent models of IBD (13). Prednisolone does prevent the complete loss of
the mucosal layer observed in the induced vehicle-treated animals
Topiramate gene expression signature
(TNBS + vehicle); however, the potent immunosuppressive effects
Fold change
of prednisolone likely slow healing from the initial chemical insult
- IL-10 signaling
and may render damaged tissues more susceptible to secondary bac-
Univ RNA normalized to 18S
terial infections.
Although the experimental results from the rodent model of IBD
Up-regulated genes
Down-regulated genes
corroborate our computational predictions derived from gene expres-
TNBS + topiramate
sion measurements of human disease, there are several caveats that
Fig. 4. Evaluation and comparison of gene expression signatures for the
could potentially limit the interpretation of the results. Foremost, we
drug compound topiramate and CD. (A) Comparison of the CD signature
have only demonstrated the efficacy of topiramate for IBD in one animal
(right) to the topiramate gene expression profile (left) showing a generally
model, which may not be representative of IBD pathology in humans.
anticorrelated pattern, with genes at the top- and bottommost ends of the
The TNBS model used in this study is the most widely used rodent
expression pattern showing generally oppositional expression patterns.
model for human IBD, and previous molecular studies have established
Genes that are up-regulated are shown in green. Genes that are down-
that it shares many molecular characteristics of human IBD, such as
regulated are shown in red. The pathways and functional groups signifi-
overproduction of interferon-g (IFN-g) mediated by T helper 1 (TH1)
cantly enriched (enrichment P < 0.05) in the top and bottom 25% of genes
T cells (14, 15). However, rat-based TNBS models tend to develop colitis
are shown on the right-hand side. (B) qPCR results showing that gene tran-
with fibrosis in the distal colon, whereas the ileum is the primary site of
scripts for TRPV1 and IFI30 have oppositional gene expression levels in thedisease condition (TNBS + vehicle) relative to the drug-treated condition
CD in human subjects (16). Furthermore, the TNBS-induced rodent
(TNBS + topiramate) in colon tissues collected from the animal validation
model is more representative of an acute IBD flare-up than the chronic
study, which corroborates their expected oppositional relationship from ini-
condition; therefore, additional studies are needed to determine whether
tial comparison of the topiramate and Crohn's expression signatures.
topiramate could serve as an effective long-term therapy for chronic IBD
MAPK3, ALOX5, and CHRM3 also showed transcript abundance trends in
in humans. Next, we noted that only two of the eight genes from the
the expected direction but did not reach statistical significance (n = 12 rats
human IBD gene signature chosen for PCR validation in the rodent
per group). *P < 0.05; **P < 0.005, two-sided Mann-Whitney U test. Error
model showed a statistically significant difference between the induced
bars, SEM. Univ RNA, Universal Rat Reference RNA.
untreated (TNBS + vehicle) and the topiramate-treated groups (TNBS +topiramate). This may be explained by species differences in gene expres-sion between human intestinal tissue, which was used to select the genes,
and is often used to treat seizures and migraines as well as depression.
and rat intestinal tissue. In addition, several of these genes may reach
Although topiramate has not previously been suggested as a therapy
distinguishing expression levels only after a longer period of treatment.
for IBD, it has been investigated for off-label use in treating obesity and
Finally, our study evaluated only a single, albeit relatively conservative,
type 2 diabetes (9, 10) and was recently shown to be effective against
dose level of topiramate; therefore, additional studies are needed to eval-
multiple sclerosis (11). Although elucidation of the precise mechanism
uate the optimal dose ranges for use of topiramate.
of action by which topiramate acts to ameliorate the induced IBD
Here, we demonstrated that computational approaches leveraging
phenotype in our study requires follow-up investigations, functional
public gene expression microarray data can be used to infer potential
enrichment analysis reveals that sets of genes related to nuclear fac-
drug therapies for IBD and offer experimental evidence that the anti-
tor kB (NF-kB) signaling, the inflammatory response, and antigen
convulsant topiramate is capable of ameliorating disease pathophysiology
presentation—all of which are relevant to the pathophysiology of
in a TNBS-induced rodent model of IBD. Because topiramate is already
IBD—are antithetically expressed in the disease and drug expression
established as a safe and effective drug for treating neurological dis-
profiles (Fig. 4). Previous work has demonstrated that inhibition of
eases in humans, and the side effect profile is generally more favorable
carbonic anhydrase IV, a known target of topiramate, enhanced recov-
than that of most drugs typically used to treat IBD (2, 17), these results
ery from colitis in a dextran sulfate sodium–induced murine model
suggest that additional clinical investigation into the use of topiramate
of colitis (12). Here, topiramate may therefore be acting at least in
for treating IBD in human subjects could be beneficial. Additionally,
part through this mechanism to ameliorate the TNBS-induced IBD
these findings support the need for future studies in which compu-
tational approaches for leveraging publicly available molecular data
Using a TNBS-induced rodent model of IBD, we demonstrated that
for drug repurposing are further developed and applied toward ad-
induced animals treated with topiramate (TNBS + topiramate) showed
ditional diseases.
Vol 3 Issue 96 96ra76
MATERIALS AND METHODS
(distal segment of the colon) were harvested for macroscopic damagescores (MDSs) and histology. The initial pilot study used eight
Computational prediction and assessment
animals per treatment group. Two independent replication studies
of novel IBD therapies
used 12 animals per treatment group. The initial animal study was
Computational prediction and assessment of novel IBD therapies was
performed at Stanford University School of Medicine. The indepen-
performed as described in a paper appearing in this issue (4). In brief,
dent replicate studies were performed by Biomodels LLC and MD
publicly available gene expression measurements were obtained from
Biosciences Inc.
NCBI GEO (6), with data from a previously published experiment mea-suring CD and UC in human intestinal tissue obtained by biopsy
Quantitative reverse transcription–PCR analysis
(GDS2642). Array probes were mapped to NCBI Entrez gene identifiers
Extraction of RNA. RNA was extracted from tissue with a standard
with the AILUN system (18). In cases where multiple microarray probes
protocol for Trizol (Invitrogen, Life Technologies)/chloroform. In brief,
mapped to the same NCBI gene identifier, we averaged across indi-
frozen colon tissues were transferred into 1 ml of Trizol and homog-
vidual probe expression values and assigned the averaged value to the
enized in a rotor stator homogenizer (Polytron PT 1200 E, Kinematica
gene. Using the significance analysis of microarrays (SAM) software, we
AG). After homogenization and addition of chloroform, tissue suspen-
created a disease gene expression signature (that is, disease signature) by
sions were centrifuged at 4°C for 15 min, resulting in the formation of
deriving the set of differentially expressed genes between the disease-
three phases. Total RNA, located in the upper aqueous phase, was sub-
affected and healthy control samples (19).
sequently precipitated with isopropanol (100%). After two wash steps
We systematically compared gene expression profiles of drugs ob-
in 75% ethanol, RNA was pelleted and diluted in tris-EDTA buffer
tained from the Connectivity Map (5) to derive a therapeutic score. A
(Ambion, Life Technologies). Concentration of RNA was assessed by
randomization approach was used to determine statistical significance,
NanoDrop (Thermo Fisher Scientific Inc.).
and drugs with predicted therapeutic scores below the significance
Reverse transcription. For complementary DNA (cDNA) syn-
threshold were ranked in reverse order according to their score. Drugs
thesis, 2 mg of total RNA was reverse-transcribed with SuperScript III
with significant negative scores have gene expression patterns that are
(Invitrogen) according to the manufacturer's protocol.
anticorrelated, or in opposition to the disease gene expression pattern,
Gene expression analyses. Transcriptional levels for nine genes,
and therefore represent putative new therapeutic indications. Specific
including 18S as an endogenous control gene, were assessed in 36 sam-
details regarding the genes and expression levels contributing to the
ples (12 controls, 12 vehicle-treated, and 12 drug-treated rats) in dupli-
predicted therapeutic match between IBD and topiramate are listed
cates with quantitative reverse transcription–PCR (qRT-PCR). All
in table S1.
reagents were purchased from Applied Biosystems, and reactions wererun on a TaqMan HT7900 machine (Applied Biosystems, Life Technol-
Functional analysis of IBD and topiramate
ogies) for a total of 40 cycles. Gene expression assays had the following
expression signatures
accession numbers: 18S (Hs99999901_s1), ALOX5 (Rn00563172_m1),
Functional analysis of the up- and down-regulated genes in the Crohn's
ALOX5AP (Rn00568506_m1), TRPV1 (Rn00583117_m1), CHRM3
and topiramate signatures (Fig. 4) was performed through the use of
(Rn00560986_s1), IFI30 (Rn01420317_m1), MAPK3 (Rn00820922_g1),
IPA (Ingenuity Systems). This functional analysis identified the
IL-6 (Rn01410330_m1), and IL-10 (Rn00563409_m1). Gene expression
biological functions and/or diseases that were most significant to the
levels were calculated with the DDCt method.
data set. Genes in the top and bottom 25% of rank change versus normalin both drug-affected and disease-affected conditions and that were as-
Macroscopic damage assessment
sociated with biological functions and/or diseases in the Ingenuity
MDS was assessed in a blinded fashion by examination of excised
Knowledge Base were considered for the analysis. Right-tailed Fisher's
colons harvested at study termination. After removal of the distal colon
exact test was used to calculate a P value determining the probability
segment ( 5 to 7 cm), tissue specimens were cut longitudinally, flushed
that each biological function and/or disease assigned to that data set
gently with normal saline to clear the fecal matter, and photographed.
is due to chance alone.
They were scored by a blinded observer for tissue damage with thefollowing criteria: 0, no inflammation; 1, swelling or redness; 2, swelling
Experimental evaluation of topiramate
and redness; 3, one or two ulcers; 4, more than two ulcers or one large
in a rodent TNBS model of IBD
ulcer; 5, necrosis.
Experimental validation of treatment efficacy was performed with aTNBS (also known as picryl sulfonic acid)–induced rodent model of
Histopathology and microscopic damage assessment
IBD (20, 21). Male Sprague-Dawley rats weighing 300 to 350 g were
A piece of the colon tissue was placed in 10% buffered formalin and
used in the study. Rats were fasted overnight before initialization of
fixed overnight. The next day, they were paraffin-embedded and cut
the study. TNBS (100 mg/kg) diluted in 50% ethyl alcohol was admin-
into 5-mm sections. The sections were stained with hematoxylin and
istered via a polyethylene catheter (PE-90) 4 to 5 cm from the anus as
eosin (H&E) and examined under light microscopy. The slides were
a rectal enema to rats under 2% isoflurane inhalation anesthesia. The
scored according to a grading system established previously (22).
control group of rats got equal volume of the vehicle (50% ethyl alcoholin water). Topiramate (80 mg/kg) dissolved in 1 M sodium hydroxide
[2.5% (v/v)] or vehicle used for topiramate was administered through
Endoscopy was performed in a blinded fashion with a small animal
a stainless steel feeding needle as oral gavage daily for 7 days starting
endoscope (Karl Storz Endoskope). To evaluate colitis severity, we an-
a day after TNBS administration. Rats were monitored daily for signs
esthetized animals with isoflurane and subjected them to video endos-
of colitis (diarrhea). On day 8, all the rats were killed and tissues
copy of the lower colon.
Vol 3 Issue 96 96ra76
SUPPLEMENTARY MATERIAL
13. T. M. Woodruff, T. V. Arumugam, I. A. Shiels, R. C. Reid, D. P. Fairlie, S. M. Taylor, A potent
human C5a receptor antagonist protects against disease pathology in a rat model of
inflammatory bowel disease. J. Immunol. 171, 5514–5520 (2003).
Table S1 caption.
14. E. Rivera, I. Flores, E. Rivera, C. B. Appleyard, Molecular profiling of a rat model of colitis:
Video descriptions.
Validation of known inflammatory genes and identification of novel disease-associated
Table S1. Details of drug and gene signatures (Excel file).
Video S1. Endoscopy of healthy controls.
targets. Inflamm. Bowel Dis. 12, 950–966 (2006).
Video S2. Endoscopy of TNBS-induced untreated.
15. F. Scheiffele, I. J. Fuss, Induction of TNBS colitis in mice. Curr. Protoc. Immunol. Chapter 15,
Unit 15.19 (2002).
Video S3. Endoscopy of TNBS-induced treated with prednisolone.
16. J. C. Hoffmann, N. N. Pawlowski, A. A. Kühl, W. Höhne, M. Zeitz, Animal models of inflammatory
Video S4. Endoscopy of TNBS-induced treated with topiramate.
bowel disease: An overview. Pathobiology 70, 121–130 (2002).
17. K. N. Roy Chengappa, L. K. Schwarzman, J. F. Hulihan, J. Xiang, N. R. Rosenthal; Clinical
REFERENCES AND NOTES
Affairs Product Support Study-168 Investigators, Adjunctive topiramate therapy in patientsreceiving a mood stabilizer for bipolar I disorder: A randomized, placebo-controlled trial.
J. Clin. Psychiatry 67, 1698–1706 (2006).
1. E. V. Loftus Jr., Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence,
18. R. Chen, L. Li, A. J. Butte, AILUN: Reannotating gene expression data automatically.
and environmental influences. Gastroenterology 126, 1504–1517 (2004).
Nat. Methods 4, 879 (2007).
2. D. C. Baumgart, W. J. Sandborn, Inflammatory bowel disease: Clinical aspects and established
19. V. G. Tusher, R. Tibshirani, G. Chu, Significance analysis of microarrays applied to the ionizing
and evolving therapies. Lancet 369, 1641–1657 (2007).
radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 (2001).
3. B. E. Sands, F. H. Anderson, C. N. Bernstein, W. Y. Chey, B. G. Feagan, R. N. Fedorak, M. A. Kamm,
J. R. Korzenik, B. A. Lashner, J. E. Onken, D. Rachmilewitz, P. Rutgeerts, G. Wild, D. C. Wolf,
20. A. R. Jurjus, N. N. Khoury, J. M. Reimund, Animal models of inflammatory bowel disease.
J. Pharmacol. Toxicol. Methods 50, 81–92 (2004).
P. A. Marsters, S. B. Travers, M. A. Blank, S. J. van Deventer, Infliximab maintenance therapy
21. S. Wirtz, C. Neufert, B. Weigmann, M. F. Neurath, Chemically induced mouse models of
for fistulizing Crohn's disease. N. Engl. J. Med. 350, 876–885 (2004).
intestinal inflammation. Nat. Protoc. 2, 541–546 (2007).
4. M. Sirota, J. T. Dudley, J. Kim, A. P. Chiang, A. A. Morgan, A. Sweet-Cordero, J. Sage, A. J. Butte,
22. H. S. Cooper, S. N. Murthy, R. S. Shah, D. J. Sedergran, Clinicopathologic study of dextran
Discovery and preclinical validation of drug indications using compendia of public gene
sulfate sodium experimental murine colitis. Lab. Invest. 69, 238–249 (1993).
expression data. Sci. Transl. Med. 3, 96ra77 (2011).
5. J. Lamb, E. D. Crawford, D. Peck, J. W. Modell, I. C. Blat, M. J. Wrobel, J. Lerner, J. P. Brunet,
23. Acknowledgments: We thank A. Skrenchuk and B. Oskotsky (Stanford University) for
A. Subramanian, K. N. Ross, M. Reich, H. Hieronymus, G. Wei, S. A. Armstrong, S. J. Haggarty,
computer support and E. Davydov, C. Do, S. Gross, and M. Schaub (Stanford University)for constructive discussion. Funding: This work was supported by Lucile Packard
P. A. Clemons, R. Wei, S. A. Carr, E. S. Lander, T. R. Golub, The Connectivity Map: Using
Foundation for Children's Health, the Hewlett Packard Foundation, National Institute
gene-expression signatures to connect small molecules, genes, and disease. Science
of General Medical Sciences (R01 GM079719), National Cancer Institute (R01 CA138256),
313, 1929–1935 (2006).
National Library of Medicine (T15 LM007033), Howard Hughes Medical Institute, and
6. T. Barrett, T. O. Suzek, D. B. Troup, S. E. Wilhite, W. C. Ngau, P. Ledoux, D. Rudnev, A. E. Lash,
Pharmaceutical Research and Manufacturers of America Foundation. Author contribu-
W. Fujibuchi, R. Edgar, NCBI GEO: Mining millions of expression profiles—Database andtools. Nucleic Acids Res. 33, D562–D566 (2005).
tions: J.T.D., M. Sirota, and A.J.B. designed the study and carried out the analysis.
M. Shenoy, M.M.S., R.K.P., S.R., and P.J.P. carried out validation experiments. A.A.M. and
7. P. M. Irving, R. B. Gearry, M. P. Sparrow, P. R. Gibson, Review article: Appropriate use of
A.P.C. assisted in the analysis. J.T.D., M. Sirota, and A.J.B. wrote the paper. Competing
corticosteroids in Crohn's disease. Aliment. Pharmacol. Ther. 26, 313–329 (2007).
interests: A.J.B. is on the scientific advisory board of NuMedii Inc. and Personalis and is
8. A. Thiry, J. M. Dogné, C. T. Supuran, B. Masereel, Anticonvulsant sulfonamides/sulfamates/
a paid consultant for Lilly, Johnson and Johnson, Genstruct, Tercica, Ansh Labs, and
sulfamides with carbonic anhydrase inhibitory activity: Drug design and mechanism of
Prevendia. J.T.D. is a consultant for NuMedii Inc. The other authors declare that they
action. Curr. Pharm. Des. 14, 661–671 (2008).
9. V. Khanna, S. Arumugam, S. Roy, S. Mittra, V. S. Bansal, Topiramate and type 2 diabetes: An
have no competing interests.
old wine in a new bottle. Expert Opin. Ther. Targets 12, 81–90 (2008).
10. K. Stenlöf, S. Rössner, F. Vercruysse, A. Kumar, M. Fitchet, L. Sjöström; OBDM-003 Study
Submitted 12 May 2011
Group, Topiramate in the treatment of obese subjects with drug-naive type 2 diabetes.
Accepted 15 July 2011
Diabetes Obes. Metab. 9, 360–368 (2007).
Published 17 August 2011
11. R. Bhat, R. Axtell, A. Mitra, M. Miranda, C. Lock, R. W. Tsien, L. Steinman, Inhibitory role for
GABA in autoimmune inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 2580–2585 (2010).
12. E. Mizoguchi, R. J. Xavier, H. C. Reinecker, H. Uchino, A. K. Bhan, D. K. Podolsky, A. Mizoguchi,
Citation: J. T. Dudley, M. Sirota, M. Shenoy, R. K. Pai, S. Roedder, A. P. Chiang, A. A. Morgan,
Colonic epithelial functional phenotype varies with type and phase of experimental colitis.
M. M. Sarwal, P. J. Pasricha, A. J. Butte, Computational repositioning of the anticonvulsant
Gastroenterology 125, 148–161 (2003).
topiramate for inflammatory bowel disease. Sci. Transl. Med. 3, 96ra76 (2011).
Vol 3 Issue 96 96ra76
Computational Repositioning of the Anticonvulsant Topiramate
for Inflammatory Bowel Disease
Joel T. Dudley, Marina Sirota, Mohan Shenoy, Reetesh K. Pai, Silke
Roedder, Annie P. Chiang, Alex A. Morgan, Minnie M. Sarwal,
Pankaj Jay Pasricha and Atul J. Butte (August 17, 2011)
Science Translational Medicine (96), 96ra76. [doi:
Greening Drug Discovery
and for drug development too. Repurposing existing,
Recycling is good for the environment
approved drugs can speed their adoption in the clinic because they can often take advantage of the existing rigorous safety testing required by the Food and Drug Administration and other regulatoryagencies. In a pair of papers, Sirota
. examined publicly available gene
expression data and determined the genes affected in 100 diseases and 164 drugs. By pairing drugs that correct abnormal gene expression in diseases, they confirm known effective drug-disease pairs andpredict new indications for already approved agents. Experimental validation that an antiulcer drug and an antiepileptic can be reused for lung cancer and inflammatory bowel disease reinforces the promise ofthis approach.
The authors scrutinized the data in Gene Expression Omnibus and identified a disease signature
for 100 diseases, which they defined as the set of mRNAs that reliably increase or decrease in patients
with that disease compared to normal individuals. They compared each of these disease signatures to each of the gene expression signatures for 164 drugs from the Connectivity Map, a collection of mRNAexpression data from cultured human cells treated with bioactive small molecules that is maintained a
the Broad Institute at Massachusetts Institute of Technology. A similarity score calculated by the
authors for every possible pair of drug and disease ranged from +1 (a perfect correlation of signatures)
1 (exactly opposite signatures). The investigators suggested that a similarity score of
predict that the drug would ameliorate the abnormalities in the disease and thus be an effective therapy.
This proved to be true for a number of drugs already on the market. The corticosteroid
prednisolone, a common treatment for Crohn's disease and ulcerative colitis, showed a strong similarityscore for these two diseases. The histone deacetylase inhibitors trichostatin A, valproic acid, and v
orinostat were predicted to work against brain tumors and other cancers (esophagus, lung, and colon),and there is experimental evidence that this is indeed the case. But in the ultimate test of method, the
authors confirmed two new predictions in animal experiments: Cimetidine, an antiulcer drug, predicted
by the authors to be effective against lung cancer, inhibited tumor cells in vitro and in vivo in mice. In
addition, the antiepileptic topiramate, predicted to improve inflammatory bowel disease by similarity
score, improved damage in colon tissue of rats treated with trinitrobenzenesulfonic acid, a model of the
disease. These two drugs are therefore good candidates for recycling to treat two diseases in need of
lung cancer and inflammatory bowel disease
and we now have a way to mine
available data for fast routes to new disease therapies.
Science Translational Medicine (print ISSN 1946-6234; online ISSN 1946-6242) is publishedweekly, except the last week in December, by the American Association for the Advancement of Science, 1200 New York Avenue, NW, Washington, DC 20005. Copyright 2016 by the AmericanAssociation for the Advancement of Science; all rights reserved. The title Science TranslationalMedicine is a registered trademark of AAAS.
The following resources related to this article are available online at http://stm.sciencemag.org.
This information is current as of February 16, 2016.
Visit the online version of this article to access the personalization andarticle tools:
The editors suggest related resources on Science's sites:
Obtain information about reproducing this article:
Science Translational Medicine (print ISSN 1946-6234; online ISSN 1946-6242) is publishedweekly, except the last week in December, by the American Association for the Advancement of Science, 1200 New York Avenue, NW, Washington, DC 20005. Copyright 2016 by the AmericanAssociation for the Advancement of Science; all rights reserved. The title Science TranslationalMedicine is a registered trademark of AAAS.
Source: http://dudleylab.org/wp-content/uploads/2016/02/Computational-repositioning-of-the-anticonvulsant-topiramate-for-inflammatory-bowel-disease..pdf
PRESS RELEASE February 23, 2016 Pharmaceutical Industry Veterans and Metabolic and Cardiovascular Disease Disease Experts join ARKAY Therapeutics' Board and Advisory Committee East Windsor, NJ — ARKAY Therapeutics, LLC announces the appointment of Alan Lewis, Ph.D., Ishwarlal Jialal, M.D., Ph.D., Martin Ogletree, Ph.D. and Clifford Davidson, Esq. as members of the Scientific advisory committee and Paul Jeffrey, M.B.A., Milton Grannatt, Ph.D. and Casey Case, Ph.D. as members of the board. "We are fortunate to have such high-caliber individuals join ARKAY's scientific advisory committee and the board. We are confident that ARKAY has the board and the advisory committee leadership to execute our business plan," said Ravi Kumar, Ph.D., founder and president of ARKAY Therapeutics. Dr. Lewis is the president, CEO and board member of DiaVacs, a Type 1 diabetes company. DiaVacs is developing products to reverse the onset of autoimmune diseases by re-inducing tolerance into the patient's immune system. Dr. Lewis currently serves as chairman of the board of Batu Biologics, a private biotechnology company, and director of a private biotechnology company, Targazyme, Inc. Dr. Lewis is a pharma industry veteran and a successful serial entrepreneur. He has served as chief executive officer and director of Medistem, Inc., Ambit Biosciences, and Novocell Inc. From January 2009 to June 2010, Dr. Lewis served as president and chief executive officer of the Juvenile Diabetes Research Foundation. Prior to joining Novocell, a company focused on stem cell therapy, starting in 2000, he was president of Celgene Signal Research, a wholly owned subsidiary of the Celgene Corporation, a pharmaceutical company. From February 1994 to August 2000, he was the president and chief executive officer of Signal Pharmaceuticals, Inc., where he guided the company to its successful acquisition by Celgene Corporation. From 1979 to 1994, Dr. Lewis held a number of positions at Wyeth-Ayerst Research and its predecessor, Wyeth Laboratories, Inc., including vice president of research at Wyeth-Ayerst Research. Dr. Lewis has published over 120 full manuscripts and has written and edited seven books. Dr. Lewis was a research associate at Yale University from 1972 to 1973. Dr. Lewis received a B.Sc. in physiology and biochemistry from Southampton University in the United Kingdom, and a Ph.D. in pharmacology from the University of Wales, in the UK. Dr. Jialal is Distinguished Professor of Pathology and Laboratory Medicine and Internal Medicine (Endocrinology, Diabetes and Metabolism), Director of the Laboratory for Atherosclerosis and Metabolic Research, Director, Special Chemistry and Toxicology, University of California Davis Medical Center, Sacramento. Dr. Jialal has published over 380 original papers and invited reviews in the areas of diabetes, atherosclerosis, lipid metabolism, nutrition and vascular biology. He has received numerous awards for his research, and has served on the editorial boards of numerous journals, including: American Journal of Clinical Nutrition, Journal of Molecular and Cellular Cardiology and Atherosclerosis. Currently, Dr. Jialal serves as section editor of the American Journal of Clinical Pathology for Clinical Chemistry and editor-in-chief of Metabolic Syndrome and Related Disorders. His major research interests are in the role of oxidative stress and inflammation in atherosclerosis, understanding the cellular dysfunction and the role of inflammation in metabolic syndrome, and understanding the pathobiology of diabetic vasculopathies. He also has a long-standing interest in hyperlipidemia
N° 10 Abril 2013 Publicación de contenido científico editada por GT Laboratorio S.R.L. Necochea 3274 Rosario Método enzimático UV para la determinación cuantitativa de lactato en suero, plasma o líquido LACTATO Liquid Plus cefalorraquídeo. El nuevo método enzimático UV GT Lab para la determinación de lactato permite la cuantificación rápida y precisa, empleando un reactivo de tra-bajo único de sencilla preparación. Dicha preparación se hace por disolución de un polvo en el buffer provis-to listo para usar. El reactivo de trabajo es estable 24 horas a temperatura ambiente o 15 días refrigerado. Se resume seguidamente el documento "Lactato: utilidad clínica y reco-mendaciones para su medición", de la Sociedad Española de Química Clí-nica preparado por P. Guevara Ramírez, R. Díaz García, A. Galán Ortega, E. Guillén Campuzano, S. Malumbres, J.L. Marín Soria, M. Muñoz Pérez, X. Navarro Segarra, P. Oliver Sáez, E. Oujo, N. del Río Barcenilla y A. Buño Soto en 2010.






