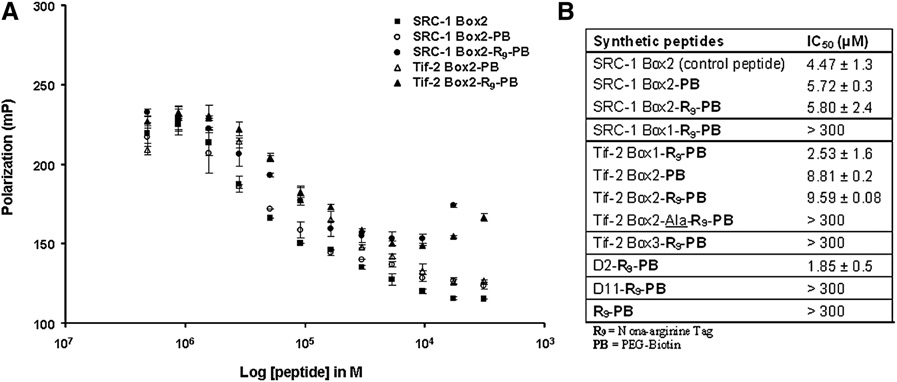
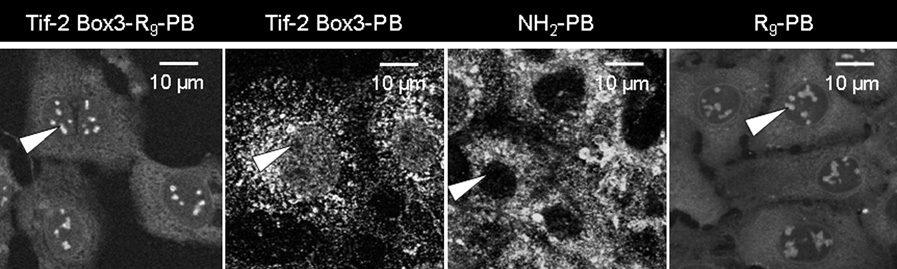
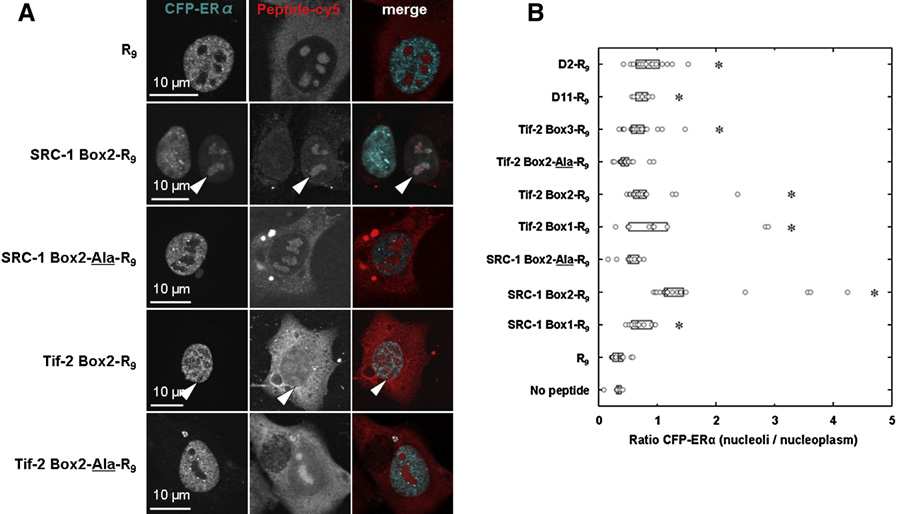
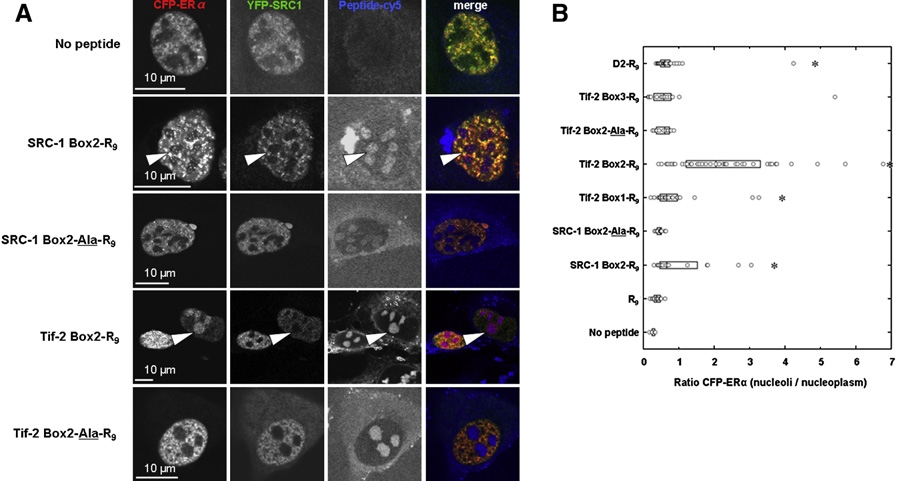
Untitled
BioFactors 25 (2005) 97–107 Coenzyme Q10 protects SHSY5Y neuronalcells from beta amyloid toxicity andoxygen-glucose deprivation by inhibiting theopening of the mitochondrial permeabilitytransition pore Geng Lia,∗, Liang-Yu Zoua,b, Chun-Mei Caoc and Edward S. Yanga aThe Jockey Club MRI Centre, The University of Hong Kong, Hong Kong, China bDepartment of Neurology, The First Affiliated Hospital, Harbin Medical University, China