Molpharm.aspetjournals.org
Copyright 2004 The American Society for Pharmacology and Experimental Therapeutics
Mol Pharmacol 66:144–152, 2004
Printed in U.S.A.
Binding of Tritiated Sildenafil, Tadalafil, or Vardenafil to thePhosphodiesterase-5 Catalytic Site Displays Potency,Specificity, Heterogeneity, and cGMP Stimulation
Mitsi A. Blount, Alfreda Beasley, Roya Zoraghi, Konjeti R. Sekhar, Emmanuel P. Bessay,Sharron H. Francis, and Jackie D. Corbin
Department of Molecular Physiology and Biophysics, Vanderbilt University School of Medicine, Nashville, Tennessee
Received January 21, 2004; accepted April 9, 2004
This article is available online at http://molpharm.aspetjournals.org
ABSTRACT
Sildenafil, tadalafil, and vardenafil each competitively inhibit
competing against one another indicated that each occupies
cGMP hydrolysis by phosphodiesterase-5 (PDE5), thereby fos-
the same site on PDE5. Studies of sildenafil and vardenafil
tering cGMP accumulation and relaxation of vascular smooth
analogs demonstrated that higher potency of vardenafil is
muscle. Biochemical potencies (affinities) of these compounds
caused by differences in its double ring. Exchange-dissociation
for PDE5 determined by IC ,
K
(isotherm),
K
studies revealed two binding components for each inhibitor.
rate), and
K (1⁄
), respectively, were the following: silde-
Excess unlabeled inhibitor did not significantly affect 3H inhib-
nafil (3.7 ⫾ 1.4, 4.8 ⫾ 0.80, 3.7 ⫾ 0.29, and 11.7 ⫾ 0.70 nM),
itor dissociation after infinite dilution, suggesting the absence
tadalafil (1.8 ⫾ 0.40, 2.4 ⫾ 0.60, 1.9 ⫾ 0.37, and 2.7 ⫾ 0.25 nM);
of subunit-subunit cooperativity. cGMP addition increased
and vardenafil (0.091 ⫾ 0.031, 0.38 ⫾ 0.07, 0.27 ⫾ 0.01, and
binding affinity of [3H]tadalafil or [3H]vardenafil, an effect pre-
0.42 ⫾ 0.10 nM). Thus, absolute potency values were similar for
sumably mediated by cGMP binding to PDE5 allosteric sites,
each inhibitor, and relative potencies were vardenafil ⬎⬎
implying that either inhibitor potentiates its own binding to
tadalafil ⬎ sildenafil. Binding of each 3H inhibitor to PDE5 was
PDE5 in intact cells by elevating cGMP. Without inhibitor
specific as determined by effects of unlabeled compounds. 3H
present, cGMP accumulation would stimulate cGMP degrada-
Inhibitors did not bind to isolated PDE5 regulatory domain.
tion, but with inhibitor present, this negative feedback process
Close correlation of EC
values using all three 3H inhibitors
would be blocked.
Phosphodiesterase-5 (PDE5) is 1 of 11 mammalian PDE
domains (
a and
b) because of their presence in cGMP-binding
families known to date (Francis et al., 2001). PDE5 is a
cyclic nucleotide PDEs, Anabaena adenylyl cyclase, and the
cGMP-specific PDE and is abundant in most smooth muscle
bacterial transcription factor FhlA (Thomas et al., 1990a;
tissues as well as in platelets, gastrointestinal epithelial
McAllister-Lucas et al., 1993; Aravind and Ponting, 1997).
cells, and Purkinje cells of the cerebellum (Francis et al.,
Isolated GAF
a monomer binds cGMP with high affinity, but
2001; Shimizu-Albergine et al., 2003). The enzyme was first
cGMP binding to GAF
b has yet to be demonstrated (Liu et
identified, purified, and cloned in this laboratory (Lincoln et
al., 2002). Allosteric binding of cGMP to PDE5 regulatory
al., 1976; Francis et al., 1980; Thomas et al., 1990a; McAllis-
domain increases affinity of the catalytic site for cGMP,
ter-Lucas et al., 1993). PDE5 is a homodimer, and each
thereby stimulating the rate of cGMP hydrolysis (Thomas et
monomer is a chimeric protein that is composed of a regula-
al., 1990b; Corbin and Francis, 1999; Okada and Asakawa,
tory domain and a catalytic domain (Corbin and Francis,
2002; Corbin et al., 2003; Mullershausen et al., 2003; Ry-
1999). The catalytic domain catalyzes the breakdown of
balkin et al., 2003). cGMP binding to the regulatory domain
cGMP to 5⬘-GMP, and the regulatory domain contains allo-
also stimulates phosphorylation of PDE5 at Ser-92 (bovine)
steric cGMP-binding sites and a phosphorylation site (Corbin
by cGMP-dependent protein kinase in vitro and in vivo
and Francis, 1999). Two tandem homologous repeats of ⬃110
(Thomas et al., 1990b; Wyatt et al., 1998; Mullershausen et
amino acids each in the regulatory domain are termed GAF
al., 2001; Murthy, 2001; Rybalkin et al., 2002). It is presumedthat cGMP binding to the regulatory domain produces a
This work was supported by National Institutes of Health Research grants
conformational change in PDE5 that exposes Ser-92. The
DK40029 and DK58277, National Institutes of Health Training grant 5T32HL-07751, and the Bayer Pharmaceuticals Corporation.
resulting phosphorylation of PDE5 increases affinity of the
ABBREVIATIONS: PDE, cyclic nucleotide phosphodiesterase; GAF, mammalian cGMP-binding phosphodiesterase,
Anabaena adenylyl cyclases,
Escherichia coli FhlA; IBMX, 3-isobutyl-1-methylxanthine; KPM, 10 mM potassium phosphate, pH 6.8, containing 15 mM -mercaptoethanol.
3H Inhibitor Binding to PDE5
regulatory domain for cGMP and increases catalytic activity
2003; Francis et al., 2003). All three 3H inhibitors were resolved in
as well (Corbin et al., 2000). These effects suggest that PDE5
single peaks and coeluted with purified unlabeled inhibitors, sug-
is critically involved in negative feedback regulation of cel-
gesting that the 3H inhibitors were unaltered after storage. Even so,
lular cGMP levels.
it cannot be completely ruled out that the curvilinearity observed in
Several compounds that potently inhibit PDE5 have been
the dissociation of 3H inhibitors in Fig. 5 could be caused by slightstructural heterogeneity of the inhibitors.
synthesized recently, and three of these are now in clinical
Isolated Regulatory Domain of PDE5. Residues Met1 to Glu539
use for treatment of male erectile dysfunction. After sexual
of human PDE5 were amplified from the hPDE5 cDNA (courtesy of
arousal, these inhibitors enhance accumulation of cGMP in
Tanabe Research Laboratories Inc., San Diego, CA). Using the forward
the smooth muscle of the arteries supplying the penis and the
sinusoids of the penile corpus cavernosum. Sildenafil (Vi-
CCCAGCT-3⬘) and the reverse primer RZGlu539rev (5⬘-GATGAT-
agra; Pfizer, New York, NY) was the first compound of this
class to be marketed for the treatment of male erectile dys-
EcoRI and NotI sites (underlined) and a stop codon (bold italic). The
function. It also shows promise in the clinical treatment of
resulting PCR fragment (1649 base pairs) was cloned into pCR 2.1-Topo
ailments related to smooth muscle tissues, such as pulmo-
(Invitrogen, Carlsbad, CA) and verified by sequencing. The fragment
nary hypertension (Weimann et al., 2000). Newer PDE5 in-
was excised by digestion with EcoRI and NotI and was inserted intobaculovirus transfer pAcHLT-A (BD PharMingen, San Diego, CA) di-
hibitors that have the same therapeutic mechanism as silde-
gested with the same enzymes. The resulting plasmid was cotrans-
nafil, such as tadalafil (Cialis; Lilly-ICOS, Bothell, WA), and
fected with the BaculoGold baculovirus DNA (BD PharMingen) into Sf9
vardenafil (Levitra; Bayer Corporation, West Haven, CT),
cells according to the manufacturer's instructions. The transfected cells
have also been approved for use in many countries. The
were incubated at 27°C for 5 days. Afterward, 100 l of collected culture
availability of these high-affinity inhibitors provides signifi-
medium was used to infect 2 ⫻ 107 freshly prepared Sf9 cells for viral
cant new tools for studies of the PDE5 catalytic domain. This
amplification. The recombinant baculovirus was amplified two more
laboratory recently examined some characteristics of the cat-
times to obtain a high titer stock solution by infecting freshly seeded Sf9
alytic domain and its regulation by investigating [3H]silde-
cells. The infected cells were incubated at 27°C for 4 days before protein
nafil binding to the enzyme (Corbin et al., 2003). The struc-
was harvested. Purification was carried out using nickel/nitrilotriacetic
tures of tadalafil and vardenafil differ significantly from that
acid agarose as described previously (Corbin et al., 2003).
of sildenafil, and these three compounds have differing in-
PDE Assays. PDE activity was determined using a modified
method (Martins et al., 1982) as described previously (Gopal et al.,
hibitory potencies. Molecular contacts of the three inhibitors
2001) with 0.4 M [3H]cGMP as substrate.
within the catalytic site of the PDE5 have recently been
[3H]cGMP-Binding Assay. The procedure was modified slightly
revealed by X-ray crystallography (Sung et al., 2003). In
from that described previously (Corbin et al., 2000). PDE5 or PDE5
addition to [3H]sildenafil, we have synthesized or acquired
(80 l) isolated regulatory domain (4 nM final protein concentration
[3H]tadalafil and [3H]vardenafil. The availability of these
in reaction mixture) was added to 2 ml of a mixture of 0.2 M
compounds has allowed a thorough analysis of the interac-
[3H]cGMP, 10 mM potassium phosphate, pH 6.8, 25 mM 2-mercap-
tion of these agents with PDE5, which is reported herein.
toethanol, and 0.2 mg/ml Type II-AS histone (Sigma). After 45 min at
These radiolabeled inhibitors have also permitted the most
4°C, the sample was filtered onto premoistened Millipore filters (pore
comprehensive, head-to-head comparison of potencies of
size, 0.45 m), which were then rinsed with 3 ml of 10 mM potassium
these agents to bind to PDE5 using several approaches.
phosphate, pH 6.8, and 25 mM -mercaptoethanol, dried, andcounted.
Moreover, some novel features of the inhibitors and of PDE5
3H Inhibitor Membrane Filtration-Binding Assay. Full-
are uncovered using these approaches.
length bovine His-tagged PDE5 (80 l) was added to 2 ml of a bindingreaction mixture that contained 0.2 mg/ml histone IIA-S, various
Materials and Methods
concentrations of 3H inhibitor, and buffer that consisted of 10 mMpotassium phosphate, pH 6.8, and 25 mM -mercaptoethanol (KPM).
Materials. [3H]cGMP and DEAE-Sephacel were purchased from
Sticking of 3H inhibitor to the sides of the test tube occurred when 3H
Amersham Biosciences Inc. (Piscataway, NJ). 3-Isobutyl-1-methyl-
inhibitor was added in the absence of or before addition of histone.
xanthine (IBMX), histone type II-AS,
Crotalus atrox snake venom,
Histone also increased retention of PDE5 on the Millipore mem-
5⬘-GMP, and cGMP were obtained from Sigma Chemical Co. (St.
branes. Binding reaction mixture containing the enzyme was incu-
Louis, MO). His-tagged, full-length recombinant bovine PDE5 was
bated on ice or in a 30°C water bath for 45 min. Millipore nitrocel-
isolated from infected Sf9 cells using nickel/nitrilotriacetic acid aga-
lulose membranes (0.45 m) were placed under house vacuum and
rose (QIAGEN, Valencia, CA) as described previously (Corbin et al.,
prewetted with 1 ml of ice-cold 10 mM potassium phosphate, pH 6.8,
2003). Native bovine lung PDE5 was obtained and purified using
that contained 0.1% Triton X-100. Next, 200 l of 25% Triton X-100
Blue Sepharose described in an earlier report (Francis and Corbin,
at room temperature in KPM was added to the reaction tube. The
1988; Thomas et al., 1990a). Sildenafil was purified from Viagra
entire contents of the tube were applied to the prewetted filter. The
tablets by following the method established previously in this labo-
reaction tube was then washed with 3 ml of cold 0.1% Triton X-100
ratory (Corbin et al., 2003). Purified sildenafil was submitted to
in 10 mM potassium phosphate, pH 6.8, and the wash was also
Amersham Biosciences for radiolabeling with tritium. Tadalafil was
applied to the filter. Filter membranes were removed, dried, and
synthesized according to Daugan (2000). After confirming the com-
transferred to 6-ml scintillation vials. Nonaqueous scintillant (5 ml)
pound structure by mass spectrometry, tadalafil was submitted to
was added to the tubes, which were then placed in a scintillation
Amersham Biosciences for radiolabeling with tritium. High-perfor-
mance liquid chromatography results from Amersham indicated that
Statistical Analyses. All values are given as mean ⫾ standard
[3H]sildenafil was ⬎98% pure, whereas the [3H]tadalafil preparation
error of mean (S.E.M.) as determined by GraphPad Prism graphics
was ⬎99% pure. Vardenafil, [3H]vardenafil, demethyl-vardenafil,
software (GraphPad Software Inc., San Diego, CA). The software
and methyl-sildenafil were provided by Bayer AG (Wuppertal, Ger-
uses the following equation: S.E.M. ⫽ standard deviation/
n1/2, where
many). All three 3H inhibitors that had been stored for more than a
standard deviation is determined as [兺(y ⫺ y
)2/(
n ⫺ 1)]1
year were subjected to Sephadex G-25 chromatography, which ad-
S.E.M. values reported fit within a 95% confidence interval, which
sorbs PDE inhibitors and provides high resolution (Corbin et al.,
quantifies the precision of the mean.
Blount et al.
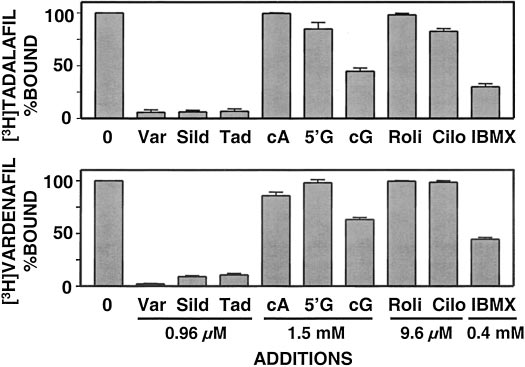
recombinant bovine PDE5 (Fig. 2). A 240-fold excess of un-labeled
Inhibition of PDE5 Catalytic Activity. The concentra-
[3H]tadalafil or [3H]vardenafil binding. Addition of cAMP or
tion of inhibitor that produces 50% inhibition of PDE5 cata-
5⬘-GMP at 375,000-fold excess did not affect binding of either
lytic activity (IC ) was determined for each of the inhibitors
inhibitor. At 375,000-fold excess, cGMP reduced binding of
(sildenafil, tadalafil, and vardenafil) using 0.4 M [3H]cGMP
either 3H inhibitor by 40 to 60%. A 2400-fold excess of rolip-
as substrate (Fig. 1). The IC
values were the following:
ram (a PDE4-specific inhibitor) or cilostamide (a PDE3-spe-
sildenafil, 3.7 ⫾ 1.4 nM (n ⫽ 4); tadalafil, 1.8 ⫾ 0.4 nM (n ⫽
cific inhibitor) did not affect 3H inhibitor binding. IBMX, a
7); and vardenafil, 0.091 ⫾ 0.031 nM (n ⫽ 5). Similar values
general, albeit weak, PDE inhibitor had a substantial inhibitory
were obtained when using native bovine PDE5 (data not
effect at 100,000-fold excess. The data suggested that binding of
shown). These values agreed with the range of published IC50
all three inhibitors is specific for the catalytic domain of PDE5
values [sildenafil, 1–9 nM (Ballard et al., 1998; Turko et al.,
and that all three inhibitors compete for the same site.
1999; Corbin and Francis, 2002); tadalafil, 1–7 nM (Corbin et
Lack of Binding of Each of the 3H Inhibitors to an
al., 2002; Gresser and Gleiter, 2002); and vardenafil, 0.1–0.8
nM (Saenz de Tejada et al., 2001; Gresser and Gleiter, 2002;
[3H]cGMP bound to the isolated regulatory domain of PDE5
Corbin et al., 2002)].
nearly stoichiometrically, none of the 3H inhibitors bound to
Stoichiometry of 3H Inhibitor Binding to PDE5. The
this domain using the same assay conditions and concentra-
binding stoichiometry was determined for each inhibitor by
tion used in the studies of binding to full-length PDE5 (data
dividing maximum binding (B
, picomoles of 3H inhibitor
not shown). Addition of a ⬃5-fold excess (0.96 M) of unla-
binding per milliliter of PDE5) obtained from GraphPad
beled sildenafil, tadalafil, or vardenafil, which was in the
Prism graphics, by PDE5 enzyme concentration (picomoles of
range of 1000 times the K of each inhibitor for the catalytic
PDE5 subunit per milliliter of PDE5). PDE5 protein concen-
domain, did not lower [3H]cGMP binding to the regulatory
tration was determined by amino acid analysis. Stoichiome-
domain (data not shown). In contrast, a 2500-fold (0.5 mM)
try was corrected for 75% recovery of 3H inhibitor binding to
excess of unlabeled cGMP, which was also approximately
PDE5 using the vacuum filtration method as determined
1000 times the K
of this ligand for the catalytic domain,
previously (Corbin et al., 2003). [3H]Tadalafil bound to PDE5
abolished [3H]cGMP binding to the regulatory domain. To-
with a stoichiometry of 0.68 ⫾ 0.10 mol/subunit (n ⫽ 7),
gether, these results indicated that inhibitor is specific for
which was similar to the [3H]vardenafil stoichiometry of
the PDE5 catalytic domain and does not bind to the regula-
0.41 ⫾ 0.05 mol/subunit (n ⫽ 8). These values compared well
tory domain under the conditions of the assays.
with the stoichiometry previously reported for [3H]sildenafil
Potencies (Affinities) for Binding of 3H Inhibitors to
of 0.61 ⫾ 0.13 mol/subunit (Corbin et al., 2003). The [3H]sil-
denafil binding stoichiometry was duplicated using the same
([3H]sildenafil, [3H]tadalafil, or [3H]vardenafil) binding to
enzyme preparation used to determine the [3H]tadalafil and
PDE5 is shown in Fig. 3. K values, obtained by using non-
[3H]vardenafil stoichiometry values calculated above.
linear regression analysis with GraphPad Prism software,
Specificity for 3H Inhibitor Binding to PDE5. The
were as follows: sildenafil, 4.8 ⫾ 0.8 nM (n ⫽ 3); tadalafil,
specificity of [3H]sildenafil binding to the catalytic domain of
2.4 ⫾ 0.6 nM (n ⫽ 4); and vardenafil, 0.38 ⫾ 0.07 nM (n ⫽ 5).
PDE5 was presented in our previous report (Corbin et al.,2003). The specificities of [3H]tadalafil and [3H]vardenafilbinding to PDE5 were determined by testing the effects ofvarious unlabeled compounds using 4 nM 3H inhibitor and
Fig. 2. Effects of nucleotides and inhibitors on binding of 3H inhibitors to
PDE5. PDE5 (0.7 nM final concentration in assay) was incubated in 2
ml of binding reaction mixture with 4 nM 3H inhibitor and the follow-
Fig. 1. Potency of inhibition of PDE catalytic activity by PDE5 inhibitors.
ing concentrations of competing compounds: unlabeled vardenafil
PDE5 (10 l; 0.113 nM final concentration in assay) was added to the
(Var) ⫽ 0.96 M, unlabeled sildenafil (Sild) ⫽ 0.96 M, unlabeled
PDE assay reaction mixture containing increasing concentrations of
tadalafil (Tad) ⫽ 0.96 M, cAMP (cA) ⫽ 1.5 mM, 5⬘-GMP (5⬘G) ⫽ 1.5
PDE5 inhibitors. PDE activity was determined in a 15-min incubation as
mM, cGMP (cG) ⫽ 1.5 mM, rolipram (Roli) ⫽ 9.6 M, cilostamide
described under Materials and Methods using 0.4 M (final concentra-
(Cilo) ⫽ 9.6 M, and IBMX ⫽ 0.4 mM. All were filtered as described
tion) [3H]cGMP as substrate. Data represent a typical experiment per-
under Materials and Methods. Data represent three experiments, each
formed in triplicate.
performed in triplicate.
3H Inhibitor Binding to PDE5
These values agreed well with the IC
Heterogeneity of the PDE5 Catalytic Domain Re-
vealed by 3H Inhibitor Dissociation Kinetics. Exchange-
Potencies for sildenafil, tadalafil, and vardenafil were also
dissociation kinetics of each of the 3H inhibitors from PDE5
determined by competition studies. For example, Fig. 4
were examined. PDE5 was first saturated with 3H inhibitor
shows the effect of increasing concentrations of unlabeled
(30 nM), and aliquots were removed to determine 3H inhibi-
vardenafil on binding of 3 nM [3H]tadalafil. The EC
tor binding at 0 time. Unlabeled inhibitor (⬃33,000-fold ex-
was calculated from GraphPad Prism graphics software us-
cess) was then added to the reaction mixture, and aliquots
ing a sigmoidal dose-response curve. Because EC
were removed for filtration at various times to follow the time
were determined using a 3H inhibitor concentration at the
course of dissociation (exchange) of the radiolabeled inhibitor
approximate K
value for PDE5, the Cheng and Prusoff/
from the enzyme. Under these conditions, the enzyme re-
Chou equation (Cheng and Prusoff, 1973; Chou, 1974) could
mained saturated at all times with inhibitor. All three inhib-
be applied to calculate the K
itors exhibited nonlinear dissociation kinetics indicative of
values by two (Table 1). It can be seen that 1⁄2 EC
the presence of at least two rate components (Fig. 5A). In Fig.
general agreement with the K or IC
for each inhibitor, and
5B, the x-axis was changed to emphasize the earlier time
the order of potency for the inhibitors was retained. The
points. Assuming the presence of two components, when the
values for unlabeled inhibitor in competition
line of the slower component was extrapolated to the y-axis,
with either [3H]vardenafil, [3H]sildenafil, or [3H]tadalafil
the calculated percentages of the two components were dif-
were similar. This suggested that the inhibitors compete for
ferent for each inhibitor. Sildenafil, as reported previously,
the same site on PDE5.
exhibited two equal components. The dissociation behavior of
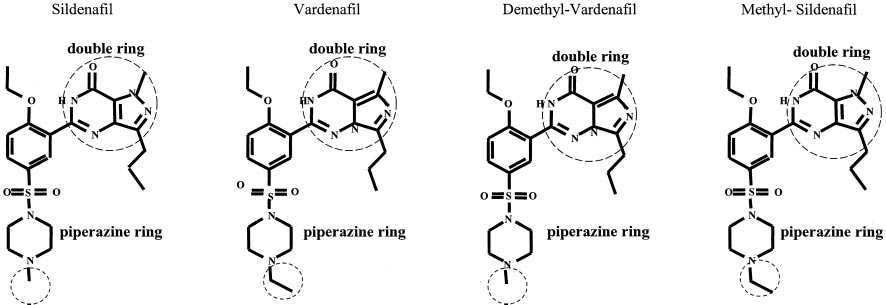
Potencies of Sildenafil and Vardenafil Analogs. Vard-
[3H]tadalafil revealed 60% high-affinity (slow) and 40% low-
enafil has a ⬃40-fold higher affinity for PDE5 over sildenafil
affinity (fast) components. [3H]Vardenafil dissociation exhib-
values shown here. To determine which of
ited 85% high-affinity and 15% low-affinity components. The
the distinguishing molecular features of the two compounds
overall rate of dissociation of [3H]vardenafil was much slower
determines this difference in potency, two analogs were
than that of the other two inhibitors. After estimation of the
synthesized. The first, demethyl-vardenafil, contained the
for dissociation, the K
of each inhibitor was calculated
[5,1-f][1,2]triazine ring of vardenafil and the appended
from the following equation: K
⫽ 6.93 ⫻ 10⫺7 M 䡠 s/t ,
methyl group of sildenafil. The second analog, methyl-
where M ⫽ molar, s ⫽ seconds, and t
sildenafil, contained the pyrazolo[4,3-d]pyrimidine ring of
seconds. (Limbird, 1995). All exchange-dissociation experi-
sildenafil and the appended ethyl group of vardenafil. The
ments were performed three times with each 3H inhibitor.
of each analog for PDE5 was determined using 0.4 M
[3H]cGMP as substrate. These experiments yielded IC50values of 0.14 ⫾ 0.02 nM for demethyl-vardenafil and
8.90 ⫾ 1.7 nM for methyl-sildenafil (Table 2). The EC
each of the analogs was determined using 0.5 nM [3H]vard-enafil. EC
values were 0.88 ⫾ 0.19 nM for demethyl-
vardenafil and 72 ⫾ 13 nM for methyl-sildenafil. K calculated
was in general agreement with the IC
analog (Table 2). The results indicated that the higher biochem-ical potency of vardenafil over sildenafil is caused by differenceswithin the double rings of the two compounds.
Fig. 4. Determination of EC
for vardenafil. Increasing concentrations of
unlabeled vardenafil were included in 2 ml of binding reaction mixturethat contained 3 nM [3H]tadalafil. PDE5 was then added (80 l; 0.035 nMfinal concentration in assay). Filtration was performed as outlined underMaterials and Methods. Data represent three experiments, each per-formed in triplicate.
values for PDE5 inhibitors
Increasing concentrations of unlabeled inhibitor were added to 2 ml of bindingreaction mixture that contained either 0.5 nM 关3H兴vardenafil, 4 nM 关3H兴sildenafil, or3 nM 关3H兴tadalafil. Filtration was performed as outlined under Materials and Meth-ods. Based on Student's t tests, the three KD values for each unlabeled inhibitor werenot significantly different from each other.
Fig. 3. Affinity of PDE5 for binding 3H inhibitors. PDE5 (80 l; 0.26 nM
final concentration in assay) was incubated with increasing concentra-
tions of 3H inhibitors in 2 ml of binding reaction mixture containing 10
M cGMP for 20 min on ice and then filtered as described under Mate-
rials and Methods. Data represent a typical experiment performed in
Blount et al.
The resulting K
values for the two [3H]sildenafil compo-
ature from 4° to 30°C had no effect or perhaps slightly inhib-
nents were 14.7 ⫾ 2.3 and 0.7 ⫾ 0.06 nM, for the two
ited sildenafil and tadalafil binding (data not shown).
[3H]tadalafil components were 9.3 ⫾ 2.67 and 0.6 ⫾ 0.00 nM,
However, the increase in temperature increased vardenafil
and for the two [3H]vardenafil components were 6.0 ⫾ 0.00
binding in the presence of cGMP, as is discussed below.
and 0.1 ⫾ 0.01 nM. The geometric mean K values for each
The effect of increasing cGMP concentrations on [3H]vard-
inhibitor (n ⫽ 3) were the following: sildenafil, 3.1 nM;
enafil binding was carried out using 0.5 nM [3H]vardenafil at
tadalafil, 1.7 nM; and vardenafil, 0.32 nM. Each average K
both 4° and 30°C (Fig. 7). At 4°C, [3H]vardenafil showed a
determined by this method was similar to IC , K obtained
2.5-fold increase in binding at low levels of cGMP (1–50 M),
from isotherm, or K
obtained from 1⁄
although this effect waned at higher cGMP concentrations.
values and average K values determined from
Repeating the experiment at 30°C with increasing cGMP
dissociation rates of the respective inhibitors suggested that
produced a ⬃3.5-fold stimulation of [3H]vardenafil binding to
interaction of the inhibitor with both kinetic components
PDE5 at 30°C. The cGMP effect remained constant at mod-
contributes to inhibition of PDE5 catalytic activity.
erate concentrations and waned slightly at very high cGMP
In addition to the exchange-dissociation method used
above, [3H]tadalafil or [3H]vardenafil dissociation from
When binding using increasing concentrations of [3H]vard-
PDE5 was examined by infinite dilution. Dissociation of the
enafil was performed at 30°C in the presence of constant 10
respective radiolabeled inhibitor was determined in the ab-
M cGMP, the labeled compound bound to PDE5 with a
sence and presence of excess unlabeled inhibitor after equi-
slightly higher affinity than at 4°C (0.42 ⫾ 0.06 nM, n ⫽ 3,
librium binding and 80-fold dilution of the binding reaction.
versus 0.59 ⫾ 0.02 nM, n ⫽ 3). [3H]Vardenafil binding to
The pattern of [3H]tadalafil dissociation (Fig. 6A) revealed
PDE5 in the absence of cGMP at 30°C yielded a lower KD
two components either in the presence or absence of a 5000-
than that found for [3H]vardenafil binding at 4°C (0.74 ⫾
fold excess of unlabeled tadalafil during dissociation. The
0.10 nM, n ⫽ 3, versus 2.19 ⫾ 0.62 nM, n ⫽ 3) (Fig. 8, A and
lack of an effect of unlabeled tadalafil on the dissociation of
[3H]tadalafil from PDE5 suggested that even though PDE5 is
The addition of 10 M cGMP to increasing concentrations
dimeric, the catalytic domain in each of the respective mono-
of [3H]vardenafil at 4°C decreased the K (0.74 ⫾ 0.10 nM, n
mers of the enzyme may not kinetically influence each other
⫽ 3, to 0.59 ⫾ 0.02 nM, n ⫽ 3) while increasing the B
to a large degree. Likewise, the dissociation of [3H]vardenafil
PDE5 (5.63 ⫾ 0.38 to 6.58 ⫾ 0.10 pmol/ml) (Fig. 8A). At 30°C,
after infinite dilution was not different from that in the
cGMP caused a 3.6-fold decrease in K from 2.19 ⫾ 0.62 nM
presence of excess vardenafil, again suggesting that the
(n ⫽ 3) to 0.42 ⫾ 0.06 nM (n ⫽ 3), whereas the B
PDE5 catalytic domains of the two monomers function inde-
significantly change (4.93 ⫾ 0.71 versus 5.25 ⫾ 0.22 pmol/ml)
pendently (Fig. 6B).
Effect of cGMP on 3H Inhibitor Binding. We recently
As shown in Fig. 9, cGMP also stimulated binding of 3 nM
reported that cGMP stimulates [3H]sildenafil binding to the
[3H]tadalafil at 4°C, and the effect was maximal at ⬃25 M
PDE5 catalytic domain at 4°C (Corbin et al., 2003). In addi-
cGMP. The stimulatory effect waned at higher cGMP concen-
tion to determining whether the same cGMP effect occurred
trations in a manner similar to the cGMP effect on vardenafil
with [3H]vardenafil and [3H]tadalafil, we also investigated if
binding at 4°C. The addition of 10 M cGMP to increasing
cGMP stimulates 3H inhibitor binding at 30°C, which ap-
concentrations of [3H]tadalafil at 4°C decreased K
proaches physiological temperature. Increasing the temper-
from 3.7 ⫾ 0.39 nM (n ⫽ 3) to 1.74 ⫾ 0.05 nM (n ⫽ 3),
values for PDE5 inhibitor analogs
Structures of analogs are shown with differences encircled. IC50 values were determined by adding PDE5 (10 l; 0.11 nM final concentration) to PDE assay reaction mixturecontaining increasing concentrations of the analogs. PDE activity was determined in a 15-min incubation as described under Materials and Methods using 0.4 M (finalconcentration) 关3H兴cGMP as substrate. EC50 values were determined by adding increasing concentrations of unlabeled inhibitor analog to 2 ml of binding reaction mixturethat contained 0.5 nM 关3H兴vardenafil. Filtration was performed as outlined under Materials and Methods. Student's t tests indicate that IC50 and KD values formethyl-sildenafil were significantly different (p ⬍ 0.05) from the IC50 and KD values for demethyl-vardenafil, vardenafil, and sildenafil.
3.7 ⫾ 1.4 (n⫽4)
0.17 ⫾ 0.04 (n⫽7)
0.14 ⫾ 0.02 (n⫽4)
8.90 ⫾ 1.7 (n⫽3)
2.47 ⫾ 0.3 (n⫽3)
1.0 ⫾ 0.26 (n⫽5)
0.44 ⫾ 0.10 (n⫽4)
36 ⫾ 6.5 (n⫽3)
3H Inhibitor Binding to PDE5
whereas the B
was 4.97 ⫾ 0.14 and 5.95 ⫾ 0.19 pmol/ml,
The isolated regulatory domain of PDE5 did not bind 3H
respectively (Fig. 10).
inhibitor using the same binding assay used for PDE5 ho-
The combined results suggested that [3H]vardenafil, but
loenzyme even though the regulatory domain bound
not [3H]sildenafil or [3H]tadalafil, binds to PDE5 with higher
[3H]cGMP nearly stoichiometrically. In addition, unlabeled
affinity at 30°C than at 4°C. The affinities of all three inhib-
sildenafil, tadalafil, or vardenafil did not compete with
itors are increased by the presence of cGMP, whereas maxi-
[3H]cGMP for binding to the regulatory domain, confirming
mum binding of each inhibitor is increased only slightly by
that these inhibitors do not bind to the regulatory domain.
values determined by binding isotherms, EC , or ex-
change-dissociation agreed with IC
of each inhibitor, again
supporting the conclusion that the PDE5-specific inhibitorsinteract exclusively with the catalytic site of PDE5. Because
[3H]Sildenafil binding to PDE5 is specific for the catalytic
cGMP-binding sites in the PDE5 regulatory and catalytic
site of PDE5 (Corbin et al., 2003). The present report dem-
domains are evolutionarily and biochemically distinct, this
onstrates that [3H]tadalafil and [3H]vardenafil are also spe-
result was not surprising.
cific for binding to the catalytic site. Binding of each of the
This laboratory has used membrane vacuum filtration to
three 3H inhibitors was inhibited by catalytic site-selective
measure [3H]cGMP binding (Francis and Corbin, 1988), 65Zn
agents and by unlabeled sildenafil, tadalafil, or vardenafil,
binding (Francis et al., 1994), and [3H]sildenafil binding
suggesting that binding of each inhibitor is restricted to the
(Corbin et al., 2003). This assay was modified slightly for
catalytic domain and that all three inhibitors also bind to the
specific [3H]tadalafil and [3H]vardenafil binding to PDE5. All
same catalytic site. The stoichiometry of each 3H inhibitor
three 3H inhibitor binding assays produced high recoveries
binding approached 1 mol/PDE5 subunit, which was consistent
and yielded nearly 1 mol/subunit binding. Radiolabeled roli-
with inhibitor binding specifically to the catalytic site and also
pram binding to PDE4 has been reported (Schneider et al.,
was indicative of one catalytic site per PDE5 monomer.
1986; Torphy et al., 1992); however, the stoichiometry of
Fig. 5. Exchange-dissociation of 3H inhibitor from PDE5. PDE5 (0.35 nM
final concentration in assay) was added to 4.5 ml of binding reaction
mixture containing 3H inhibitor (30 nM final concentration). Then, to
Fig. 6. Dissociation of 3H inhibitors from PDE5 after infinite dilution.
determine the zero time point, a 520-l aliquot of this mixture was
PDE5 (76 l, 0.32 nM final concentration in assay) was added to 360 l
filtered as described under Materials and Methods. Next, 30 l of a 1 mM
of binding reaction mixture containing a final concentration of 2 nM
solution of the corresponding unlabeled inhibitor was added to the re-
[3H]tadalafil (A) or 0.5 nM [3H]vardenafil (B). After incubating for 1 h on
maining incubating binding reaction mixture at 4°C. Aliquots were re-
ice, 35 ml of 0.2 mg/ml histone AII-S in the absence or presence of 10 M
moved and filtered by the same procedure at the indicated time points. A
of the respective unlabeled inhibitor was added to dilute the binding
represents a longer time course, whereas B shows a shorter time course
reaction mixture 80-fold. Filtration was performed at the indicated time
to emphasize curvilinear kinetics. Data represent three experiments,
points by the procedure outlined under Materials and Methods. Data
each performed in triplicate.
represent three experiments, each performed in triplicate.
Blount et al.
binding in those studies was less than 0.01 mol/subunit using
which of these structural differences of the compounds deter-
mines potency, two analogs were synthesized: demethyl-
values of sildenafil, tadalafil, and vardenafil deter-
vardenafil (analog of vardenafil containing the appended
mined here in head-to-head assays using bovine PDE5 were
methyl group of sildenafil) and methyl-sildenafil (analog of
in the same range as IC
values reported in the literature
sildenafil containing the appended ethyl group of vardenafil).
using human PDE5 (Table 3) (Corbin and Francis, 2002;
Demethyl-vardenafil and vardenafil had almost identical
Corbin et al., 2002). Therefore, results are similar using
values, whereas methyl-sildenafil had 52-times higher
recombinant PDE5, native PDE5, or PDE5 from different
which was similar to the IC
of sildenafil. K
mammalian species. Whereas IC
is the classic method of
experiments using both analogs also
determining potency (affinity) of PDE inhibitors, measure-
indicated that methyl-sildenafil had much lower potency
ment of binding strength, or K , is a more direct method of
than either of the other two analogs. From these results, the
determining potency and it also provides a measure of stoi-
higher biochemical potency of vardenafil compared with sil-
chiometry of ligand binding. This report determined the po-
denafil is caused by differences within the double rings of the
tencies for sildenafil, tadalafil, and vardenafil using four
two compounds. The crystal structure of the PDE5 catalytic
separate head-to-head methods. IC
measurements yielded
domain containing either sildenafil or vardenafil was re-
a potency ratio of 1:2:41, K
(binding isotherm) yielded a
ported recently (Sung et al., 2003); however, the resolution of
ratio of 1:2:13, K (1⁄
) yielded a ratio of 1:5:26, and K
the crystal structure was not sufficient to identify distinct
(exchange-dissociation) yielded a ratio of 1:2:14 for sildenafil,
interactions of either of these two inhibitors with the enzyme.
tadalafil, and vardenafil, respectively (Table 3). This inves-
The difference in the double ring of vardenafil, compared
tigation represents the most comprehensive examination of
with sildenafil, may possibly allow for a stronger interaction
the absolute and relative potencies of these drugs.
between the compound and one or more of the amino acids
Dissociation rates of inhibitors from PDE5 correlated with
(e.g., Tyr-612, Val-782, Phe-820, Leu-785, and Gln-817) that
potencies determined by IC
or isotherm K , i.e., the slower
the rate, the higher the potency. However, the faster disso-ciation rate of tadalafil from PDE5 compared with that ofvardenafil may be unexpected in view of the longer lastingclinical effects of tadalafil. These clinical differences oftadalafil may be caused by pharmacokinetic considerationssuch as slower intestinal absorption or slower degradation bythe liver, rather than by different biochemical properties.
In comparing the distinctive chemical structures of silde-
nafil and vardenafil, two major differences are evident: 1) a
methyl group is appended to the piperazine ring of sildenafil,whereas the same ring in vardenafil has an appended ethylgroup, and 2) a nitrogen atom is present in the 7-position ofthe double ring of sildenafil, but it is not present in the ringof vardenafil, although vardenafil contains a nitrogen atomin the 5-position, which is absent in sildenafil. To resolve
Fig. 8. Effect of temperature on cGMP stimulation of [3H]vardenafil
binding using varying concentrations of [3H]vardenafil. PDE5 (80 l, 0.07
Fig. 7. Effect of cGMP on [3H]vardenafil binding at 4° and 30°C. PDE5
nM final concentration in assay) was added to 2 ml of binding reaction
(80 l, 0.07 nM final concentration in assay) was added to 2 ml of binding
mixture containing 0.05 to 6.4 nM [3H]vardenafil in the absence and
reaction mixture containing 0.5 nM [3H]vardenafil and 0 to 350 M
presence of 10 M cGMP and incubated for 45 min on ice (A) or for 20 min
cGMP and incubated for 45 min on ice or 20 min in a 30°C water bath.
in a 30°C water bath (B). Binding was performed as outlined under
Filtration was performed as outlined under Materials and Methods.
Materials and Methods. Units indicate picomoles of inhibitor per millili-
Units indicate picomoles of inhibitor per milliliter. Data represent three
ter of PDE5 added to the reaction. Data represent three experiments,
experiments, each performed in triplicate.
each performed in triplicate.
3H Inhibitor Binding to PDE5
could be important for binding of the double ring of the
sildenafil or that provides an indirect contact resulting from
inhibitor to human PDE5 (Sung et al., 2003). In addition, the
change in the electron distribution in the double ring.
position of the nitrogen atom in the vardenafil double ring
Exchange-dissociation experiments using each of the three
may impart a change in an atom or group of this molecule
3H inhibitors revealed curvilinear dissociation kinetics, sug-
that provides contact with a residue that is not contacted by
gesting the presence of two or more catalytic site compo-nents. There was an apparent link between inhibitor potencyand percentage of high-affinity (slow) component of binding.
This could mean that 1) the three 3H inhibitors selecteddifferently for binding to two preexisting populations ofPDE5 having different affinities; 2) the inhibitors had differ-ent potencies for promoting conversion of one population intoanother; or 3) a combination of both mechanisms. Dissocia-tion of 3H inhibitor induced by infinite dilution also displayedheterogeneous kinetics. One possible explanation for thepresence of two or more components of 3H inhibitor dissoci-ation is that PDE5 exists in different conformations (Franciset al., 1998). PDE2 (Manganiello et al., 1990) and PDE4
(Laliberte et al., 2000) also demonstrated kinetic heteroge-neity, which was interpreted to represent different enzymeconformations. The present report is the first to extensivelydemonstrate that the PDE5 catalytic site exhibits more thanone kinetic state, but whether or not it was caused by the
Fig. 9. Effect of cGMP on [3H]tadalafil binding. PDE5 (80 l, 0.07 nM
presence of different PDE5 conformations remains to be
final concentration) was added to 2 ml of binding reaction mixture con-
proved. It cannot be ruled out that PDE5 undergoes partial
taining 3 nM [3H]tadalafil and 0 to 350 M cGMP. The mixtures were
modification during preparation, which could explain the
then incubated for 45 min on ice. Filtration was performed as outlinedunder Materials and Methods. Units indicate picomoles of inhibitor per
heterogeneity observed, although the presence of two compo-
milliliter of PDE5 added to the reaction. Data represent three experi-
nents is observed in different preparations of recombinant
ments, each performed in triplicate.
PDE5 and native PDE5. Regardless, caution must now beused in interpreting binding isotherm K values that assume
the presence of a single component in the calculation (Corbinet al., 2003).
Cooperativity of inhibitor binding to PDE5 might occur if
binding of inhibitors to the catalytic site of one of the twosubunits affects binding to the other subunit. However, in-hibitor dissociation after infinite dilution in the absence andpresence of excess unlabeled inhibitor indicated that this isnot the case, at least under the conditions used for the ex-periment.
Whereas the molecular mechanism for stimulation of
PDE5 catalytic activity by cGMP binding to the regulatorydomain is unknown, it is suggested that cGMP binding tothis domain relieves PDE5 of an autoinhibitory constraint, atwhich point filling of the catalytic site at subsaturating sub-strate levels of cGMP is facilitated, increasing catalytic ac-tivity. This negative feedback mechanism promotes rapid
Fig. 10. Effect of cGMP on [3H]tadalafil binding using varying concen-
degradation of cGMP within the cell. This negative feedback
trations of [3H]tadalafil. PDE5 (80 l, 0.07 nM final concentration inassay) was added to 2 ml of binding reaction mixture containing 0.05 to
could be problematic for individuals with erectile dysfunction
6.4 nM [3H]tadalafil in the absence and presence of 10 M cGMP, and the
who are unable to maintain the high level of cGMP in the
mixture was incubated for 45 min on ice. Filtration was performed as
corpus cavernosum for the extended time that is required to
outlined under Materials and Methods. Units indicate picomoles of in-
achieve and maintain penile erection. This potential defi-
hibitor per milliliter of PDE5 added to the reaction. Data represent threeexperiments, each performed in triplicate.
ciency is apparently overcome by the presence of nonhydro-
TABLE 3Head-to-head comparison of PDE5-specific inhibitor potencies (affinities)Student's t tests indicated that KD values for each unlabeled inhibitors were significantly different from each other with the exception of the KD value of sildenafil obtainedfrom 1/2 EC50, which was significantly different (p ⬍ 0.05) from all other KD and IC50 values for sildenafil. The IC50 value for vardenafil was significantly different (p ⬍ 0.05)from all KD values for vardenafil.
D from Exchange-Dissociation Average
IC50 from Literature
Blount et al.
lyzable PDE5 inhibitors that are specific for the catalytic site.
Francis SH, Sekhar KR, Rouse AB, Grimes KA, and Corbin JD (2003) Single step
isolation of sildenafil from commercially available Viagra tablets. Int J Impot Res
The inhibitors may increase cGMP levels by blocking the
negative feedback process while simultaneously increasing
Francis SH, Turko IV, and Corbin JD (2001) Cyclic nucleotide phosphodiesterases:
cGMP levels by competition.
relating structure and function. Prog Nucleic Acid Res Mol Biol 65:1–52.
Gopal VK, Francis SH, and Corbin JD (2001) Allosteric sites of phosphodiesterase-5
Because cGMP stimulates binding of [3H]tadalafil and
(PDE5). A potential role in negative feedback regulation of cGMP signaling in
[3H]vardenafil, as well as [3H]sildenafil, to the PDE5 cata-
corpus cavernosum. Eur J Biochem 268:3304 –3312.
Gresser U and Gleiter CH (2002) Erectile dysfunction: comparison of efficacy and
lytic site, cGMP stimulation is not inhibitor-specific. Thus,
side effects of the PDE-5 inhibitors sildenafil, vardenafil and tadalafil—review of
cGMP stimulation should also lower the level of drug that
the literature. Eur J Med Res 7:435– 446.
Laliberte F, Han Y, Govindarajan A, Giroux A, Liu S, Bobechko B, Lario P, Bartlett
can be administered to cause smooth cell relaxation, which is
A, Gorseth E, Gresser M, et al. (2000) Conformational difference between PDE4
desirable to minimize side effects and safety concerns.
apoenzyme and holoenzyme. Biochemistry 39:6449 – 6458.
Lincoln TM, Hall CL, Park CR, and Corbin JD (1976) Guanosine 3⬘:5⬘-cyclic mono-
A relatively high concentration (1–25 M) of cGMP was
phosphate binding proteins in rat tissues. Proc Natl Acad Sci USA 73:2559 –2563.
required to stimulate maximal binding of both [3H]tadalafil
Limbird LE (1995) Cell Surface Receptors: A Short Course on Theory and Methods,
and [3H]vardenafil. This concentration was unusually high
2nd ed, Kluwer Academic Publishers, Boston.
Liu L, Underwood T, Li H, Pamukcu R, and Thompson WJ (2002) Specific cGMP
considering that previous results indicated that the K
binding by the cGMP binding domains of cGMP-binding cGMP specific phospho-
cGMP binding to the GAF domains is 0.2 M (Thomas et al.,
diesterase. Cell Signal 14:45–51.
Manganiello VC, Smith CJ, Degerman E, and Belfrage P (1990) Cyclic GMP-
1990b). The apparent discrepancy could be caused by the
inhibited cyclic nucleotide phosphodiesterases, in Cyclic Nucleotide Phosphodies-
different assay conditions used for measuring binding affin-
terases: Structure, Regulation and Drug Action (Beavo J and Houslay M eds)
pp 87–109, John Wiley and Sons, New York.
ities of [3H]cGMP and 3H inhibitors. On the other hand,
Martins TJ, Mumby MC, and Beavo JA (1982) Purification and characterization of a
although it has been shown so far that cGMP binds only to a
cyclic GMP-stimulated cyclic nucleotide phosphodiesterase from bovine tissues.
J Biol Chem 257:1973–1979.
high-affinity GAF a site (Liu et al., 2002), the high concen-
McAllister-Lucas LM, Sonnenburg WK, Kadlecek A, Seger D, LeTrong H, Colbran
trations of cGMP required to stimulate 3H inhibitor binding
JL, Thomas MK, Walsh KA, Francis SH, Corbin JD, et al. (1993) The structure ofa bovine lung cGMP-binding, cGMP-specific phosphodiesterase deduced from a
to PDE5 suggests additional binding to a lower affinity GAF
cDNA clone. J Biol Chem 268:22863–22873.
b site, which could lead to increased catalytic site affinity for
Mullershausen F, Friebe A, Feil R, Thompson WJ, Hofmann F, and Koesling D
(2003) Direct activation of PDE5 by cGMP: long-term effects within NO/cGMP
signaling. J Cell Biol 160:719 –727.
Mullershausen F, Russwurm M, Thompson WJ, Liu L, Koesling D, and Friebe A
(2001) Rapid nitric oxide-induced desensitization of the cGMP response is causedby increased activity of phosphodiesterase type 5 paralleled by phosphorylation of
We thank Dr. David Wood for excellent advice during preparation
the enzyme. J Cell Biol 155:271–278.
Murthy KS (2001) Activation of phosphodiesterase 5 and inhibition of guanylate
of this manuscript. We are grateful to Bayer for providing PDE5
cyclase by cGMP-dependent protein kinase in smooth muscle. Biochem J 360:199 –
inhibitor analogs. We thank Eric Howard of the Vanderbilt Univer-
sity Protein Chemistry Core for amino acid analyses. We are also
Okada D and Asakawa S (2002) Allosteric activation of cGMP-specific, cGMP-
binding phosphodiesterase (PDE5) by cGMP. Biochemistry 41:9672–9679.
grateful to the E. Bronson Ingram Cancer Center and Diabetes
Rybalkin SD, Rybalkina IG, Feil R, Hofmann F, and Beavo JA (2002) Regulation of
Center of Vanderbilt University.
cGMP-specific phosphodiesterase (PDE5) phosphorylation in smooth muscle cells.
J Biol Chem 277:3310 –3317.
Rybalkin SD, Rybalkina IG, Shimizu-Albergine M, Tang XB, and Beavo JA (2003)
PDE5 is converted to an activated state upon cGMP binding to the GAF A domain.
Aravind L and Ponting CP (1997) The GAF domain: an evolutionary link between
EMBO (Eur Mol Biol Organ) J 22:469 – 478.
diverse phototransducing proteins. Trends Biochem Sci 22:458 – 459.
Saenz de Tejada I, Angulo J, Cuevas P, Fernandez A, Moncada I, Allona A, Lledo E,
Ballard SA, Gingell CJ, Tang K, Turner LA, Price ME, and Naylor AM (1998) Effects
Korschen HG, Niewohner U, Haning H, et al. (2001) The phosphodiesterase
of sildenafil on the relaxation of human corpus cavernosum tissue in vitro and on
inhibitory selectivity and the in vitro and in vivo potency of the new PDE5
the activities of cyclic nucleotide phosphodiesterase isozymes. J Urol 159:2164 –
inhibitor vardenafil. Int J Impot Res 13:282–290.
Schneider HH, Schmiechen R, Brezinski M, and Seidler J (1986) Stereospecific
Cheng Y and Prusoff WH (1973) Relationship between the inhibition constant (K1)
binding of the antidepressant rolipram to brain protein structures. Eur J Phar-
and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an
enzymatic reaction. Biochem Pharmacol 22:3099 –3108.
Shimizu-Albergine M, Rybalkin SD, Rybalkina IG, Feil R, Wolfsgruber W, Hofmann
Chou T (1974) Relationships between inhibition constants and fractional inhibition
F, and Beavo JA (2003) Individual cerebellar Purkinje cells express different
in enzyme-catalyzed reactions with different numbers of reactants, different reac-
cGMP phosphodiesterases (PDEs): in vivo phosphorylation of cGMP-specific PDE
tion mechanisms and different types and mechanisms of inhibition. Mol Pharma-
col 10:235–247.
(PDE5) as an indicator of cGMP-dependent protein kinase (PKG) activation.
Corbin JD, Blount MA, Weeks JL 2nd, Beasley A, Kuhn KP, Ho YS, Saidi LF, Hurley
J Neurosci 23:6452– 6459.
JH, Kotera J, and Francis SH (2003) [3H]Sildenafil binding to phosphodiesterase-5
Sung BJ, Yeon Hwang K, Ho Jeon Y, Lee JI, Heo YS, Hwan Kim J, Moon J, Min Yoon
is specific, kinetically heterogeneous and stimulated by cGMP. Mol Pharmacol
J, Hyun YL, Kim E, et al. (2003) Structure of the catalytic domain of human
phosphodiesterase 5 with bound drug molecules. Nature (Lond) 425:98 –102.
Corbin JD and Francis SH (1999) Cyclic GMP phosphodiesterase-5: target of silde-
Thomas MK, Francis SH, and Corbin JD (1990a) Characterization of a purified
nafil. J Biol Chem 274:13729 –13732.
bovine lung cGMP-binding cGMP phosphodiesterase. J Biol Chem 265:14964 –
Corbin JD and Francis SH (2002) Pharmacology of phosphodiesterase-5 inhibitors.
Int J Clin Pract 56:453– 459.
Thomas MK, Francis SH, and Corbin JD (1990b) Substrate- and kinase-directed
Corbin JD, Francis SH, and Webb DJ (2002) Phosphodiesterase type 5 as a phar-
regulation of phosphorylation of a cGMP-binding phosphodiesterase by cGMP.
macologic target in erectile dysfunction. Urology 60:4 –11.
J Biol Chem 265:14971–14978.
Corbin JD, Turko IV, Beasley A, and Francis SH (2000) Phosphorylation of phos-
Torphy TJ, Livi GP, Balcarek JM, White JR, Chilton FH, and Undem BJ (1992)
phodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic
Therapeutic potential of isozyme-selective phosphodiesterase inhibitors in the
and allosteric cGMP-binding activities. Eur J Biochem 267:2760 –2767.
treatment of asthma. Adv Second Messenger Phosphoprotein Res 25:289 –305.
Daugan AC-M (2000), inventor, ICOS Corporation, assignee. Use of cGMP-
Turko IV, Ballard SA, Francis SH, and Corbin JD (1999) Inhibition of cyclic GMP-
phosphodiesterase inhibitors in methods and compositions to treat impotence. U.S.
binding cyclic GMP-specific phosphodiesterase (Type 5) by sildenafil and related
patent 6,140,329. 2000 Oct 31.
compounds. Mol Pharmacol 56:124 –130.
Francis SH, Colbran JL, McAllister-Lucas LM, and Corbin JD (1994) Zinc interac-
Weimann J, Ullrich R, Hromi J, Fujino Y, Clark MW, Bloch KD, and Zapol WM
tions and conserved motifs of the cGMP-binding cGMP- specific phosphodiesterase
(2000) Sildenafil is a pulmonary vasodilator in awake lambs with acute pulmonary
suggest that it is a zinc hydrolase. J Biol Chem 269:22477–22480.
Francis SH and Corbin JD (1988) Purification of cGMP-binding protein phosphodi-
Wyatt TA, Naftilan AJ, Francis SH, and Corbin JD (1998) ANF elicits phosphory-
esterase from rat lung. Methods Enzymol 159:722–729.
lation of the cGMP phosphodiesterase in vascular smooth muscle cells. Am J
Francis SH, Chu DM, Thomas MK, Beasley A, Grimes K, Busch JL, Turko IV, Haik
Physiol 274:H448 –H455.
TL, and Corbin JD (1998) Ligand-induced conformational changes in cyclic nucle-otide phosphodiesterases and cyclic nucleotide-dependent protein kinases. Meth-
Address correspondence to: Dr. Jackie D. Corbin, 702 Light Hall, Department of
Molecular Physiology and Biophysics, Vanderbilt University School of Medicine,
Francis SH, Lincoln TM, and Corbin JD (1980) Characterization of a novel cGMP
Nashville, TN 37232-0615. E-mail: [email protected]
binding protein from rat lung. J Biol Chem 255:620 – 626.
Source: http://molpharm.aspetjournals.org/content/66/1/144.full.pdf
Breastfeeding is Best! It seems that every year in the summer just before remember being quite shocked to see an old picture of Canadians celebrate World Breastfeeding Week there one of my children bottle feeding when I ‘remembered' is media coverage of something that undermines him as exclusively breastfed. breastfeeding. This year we seem to have gotten an
Tips for Knowing Your LEVOXYL Take note of your specific LEVOXYL dose LEVOXYL tablets can be identified by their unique thyroid shape and are color coded by strength. Print and fill out this form to take to the pharmacy so you can make sure you get the brand your doctor has prescribed. My LEVOXYL dosage(s) strength(s): My LEVOXYL tablet color(s): Circle the dosage(s) strength(s) you've been prescribed and check to make sure you have received the correct strengths and colors at the pharmacy.



